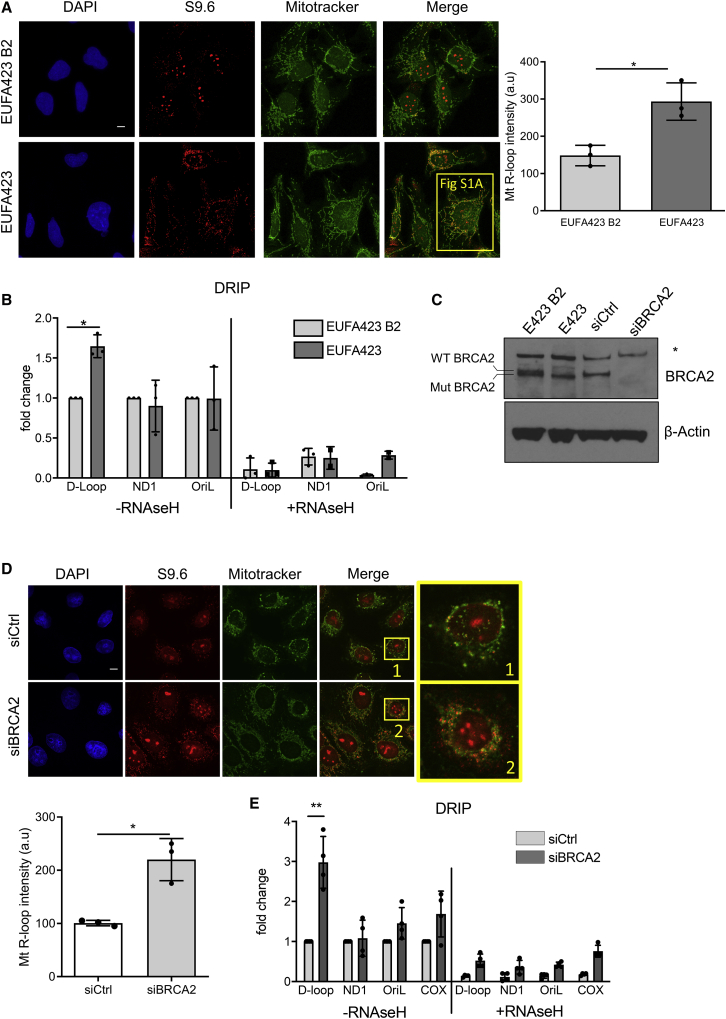
Figure 1.
R-loops accumulate in the D-loop region of mtDNA in cells lacking BRCA2
(A) Immunofluorescence detection of R-loops with S9.6 antibody in BRCA2-deficient EUFA423 cells or EUFA423-B2 controls complemented with wild-type BRCA2. Plot shows the mean ± SD from three independent experiments. At least 70 cells were counted in each condition. The two-tailed Student’s t test was performed to determine statistical significance between the two groups. ∗p < 0.05. Scale bars, 10 μm; DAPI (4,6-diamidino-2-phenylindole) stains DNA, MitoTracker stains mitochondria. Magnification of one cell indicated by a yellow square is shown in Figure S1A.
(B) DRIP analysis with S9.6 antibody in EUFA423-B2 and EUFA423 cells. RNaseH1 treatment serves as a control for antibody specificity. Plots depict the mean ± SD from three independent experiments. The two-way ANOVA test was performed for all pairs to determine statistical significance. Statistically significant differences are indicated by ∗p < 0.05.
(C) Western blot showing BRCA2 depletion using specific (si)RNA (siBRCA2) for 72 h, compared to control (siCtrl). β-actin is the loading control.
(D) Immunofluorescence detection of R-loops with S9.6 antibody in HeLa Kyoto cells treated either with siCtrl or siBRCA2. Plot shows the mean ± SD from three independent experiments. At least 70 cells were counted in each condition. The two-tailed Student’s t test was performed to determine statistical significance between the two groups. ∗p < 0.05. Scale bars, 10 μm.
(E) DRIP analysis in HeLa Kyoto cells transfected either with siCtrl or siBRCA2. DRIP was performed 72 h after transfection and depicted as described in the preceding panels. Plots depict the mean ± SD from three independent experiments. Statistically significant differences are indicated ∗∗p < 0.01.