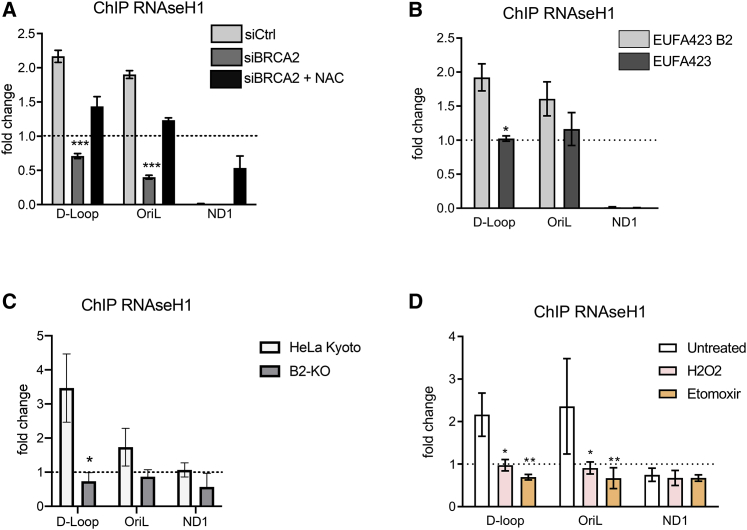
Figure 5.
Impaired recruitment of RNaseH1 to sites of R-loop formation in mtDNA
(A–D) Chromatin immunoprecipitation (ChIP) analysis with anti-RNaseH1 antibody of the mitochondrial D-loop region; OriL or ND1 gene shows similar ChIP analyses in BRCA2-KO cells compared to the parental HeLa Kyoto cells.
(B) Similar ChIP analyses in BRCA2-deficient EUFA423 cells compared to EUFA423-B2 controls expressing wild-type BRCA2.
(A and C) HeLa Kyoto cells treated either with (si)Ctrl (C) or (si)BRCA2 (A) and with 2 mM NAC for 16 h.
(D) Similar analysis of HeLa Kyto cells treated with 1 mM H2O2 or with 50 μM etomoxir for 16 h. The fold change in ChIP signal relative to an immunoglobulin G (IgG) isotype control is plotted. Plots show the mean ± SD from three independent experiments. The two-way ANOVA test was performed to determine statistical significance. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.