FIGURE 3.
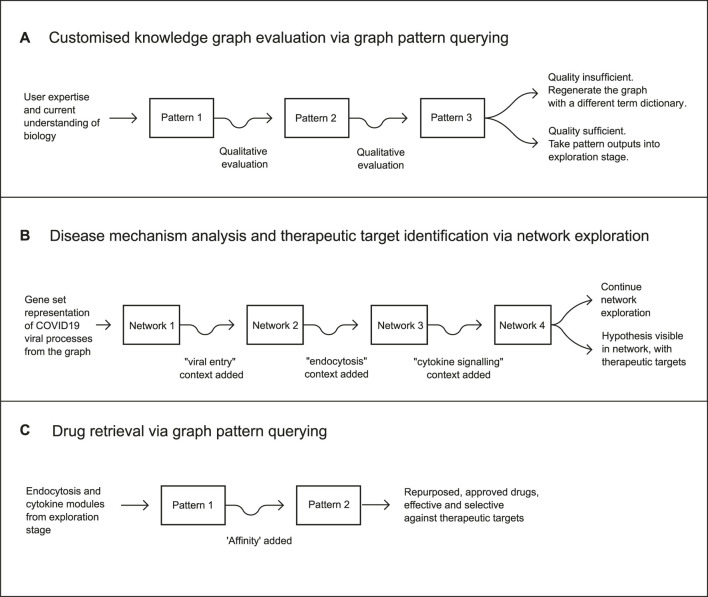
The full drug repurposing workflow consisted of three stages. The first stage incorporated the evaluation of a customised knowledge graph via graph pattern querying as a means of rapid qualitative evaluation. The second was a mechanistic analysis of viral and host processes, via the creation of an initial network using a gene set representation of COVID19-related processes extracted from the customised knowledge graph. This was iterated three more times to achieve a final network containing a clear hypothesis and therapeutic targets. The third stage resulted in a hypothesis as a partially defined graph pattern, from which targets and drug candidates were retrieved, producing a final set of candidate treatments for COVID-19.