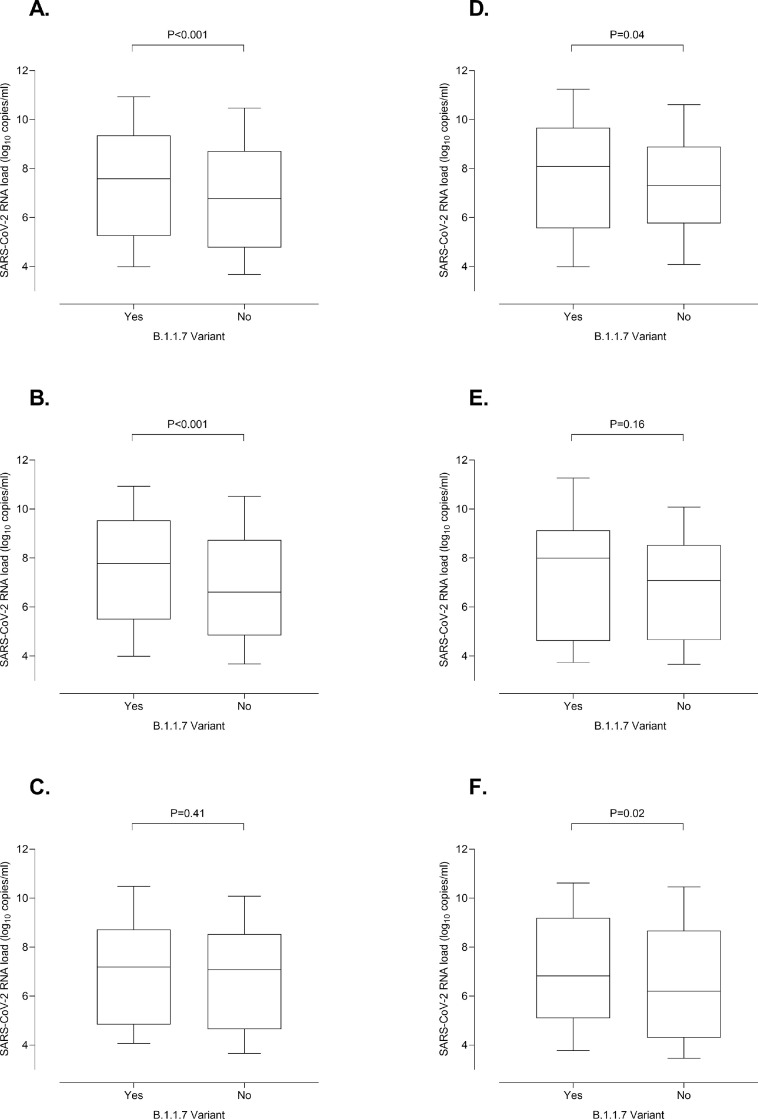
Fig. 1.
Whisker plots depicting initial SARS-CoV-2 RNA loads in nasopharyngeal specimens from individuals infected by either the B.1.1.7 variant or other variants. Participants were sampled at either primary care centers affiliated to the Clínico-Malvarrosa Health Department, which attends 350,000 inhabitants in Northwest Valencia (Spain), or at its tertiary reference hospital (Hospital Clínico Universitario de Valencia, Spain-HCU-). NP were collected by trained nurses, placed in 3 ml of Universal Transport Medium (UTM, Becton Dickinson, Sparks, MD, USA) and delivered to the Microbiology Service of HCU for testing. Specimens were analyzed by RT-PCR within 24 h of receipt. We used the TaqPath COVID-19 Combo Kit (Thermo Fisher Scientific, MS, USA), which targets SARS-CoV-2 ORF1ab, N and S genes, following RNA extraction carried out by using the Applied Biosystems™ MagMAX™ Viral/Pathogen II Nucleic Acid Isolation Kits coupled with Thermo Scientific™ KingFisher Flex automated instrument. The AMPLIRUN® TOTAL SARS-CoV-2 RNA Control (Vircell SA, Granada, Spain) was used as the reference material for estimating SARS-CoV-2 RNA load (in copies/mL, taking RT-PCR CT for the N gene). The B.1.1.7 lineage was confirmed by whole-genome sequencing in 108 cases, whereas in the remaining 230 it was inferred by S-gene target failure (SGTF) in the RT-PCR as within the timeframe of specimen collection (mid-February-April 2021) more than 95% of SGTFs detected in the Clínico-Malvarrosa Health Department belonged to that lineage (not shown). Control individuals were sampled before the SARS-CoV-2 B1.1.7 variant was first detected in our Health Department in early January 2021. Moreover, the SARS-CoV-2 RNA S-gene was detected in NP from all these participants. The data are depicted for all participants (A); adults (B), children (≤18 years old) (C); adults with COVID-19 (D); children with COVID-19 (E) and asymptomatic adults (F). The absence of non-B.1.1.7-infected asymptomatic children in the cohort precluded meaningful comparison of viral loads between these individuals and those infected by other variants. Differences between medians were compared using the Mann-Whitney U test. The Chi-squared test was used for frequency comparisons. Two-sided P-values < 0.05 were considered statistically significant. Statistical analyses were performed using the SPSS v.25 program. P values for comparisons across groups are shown.