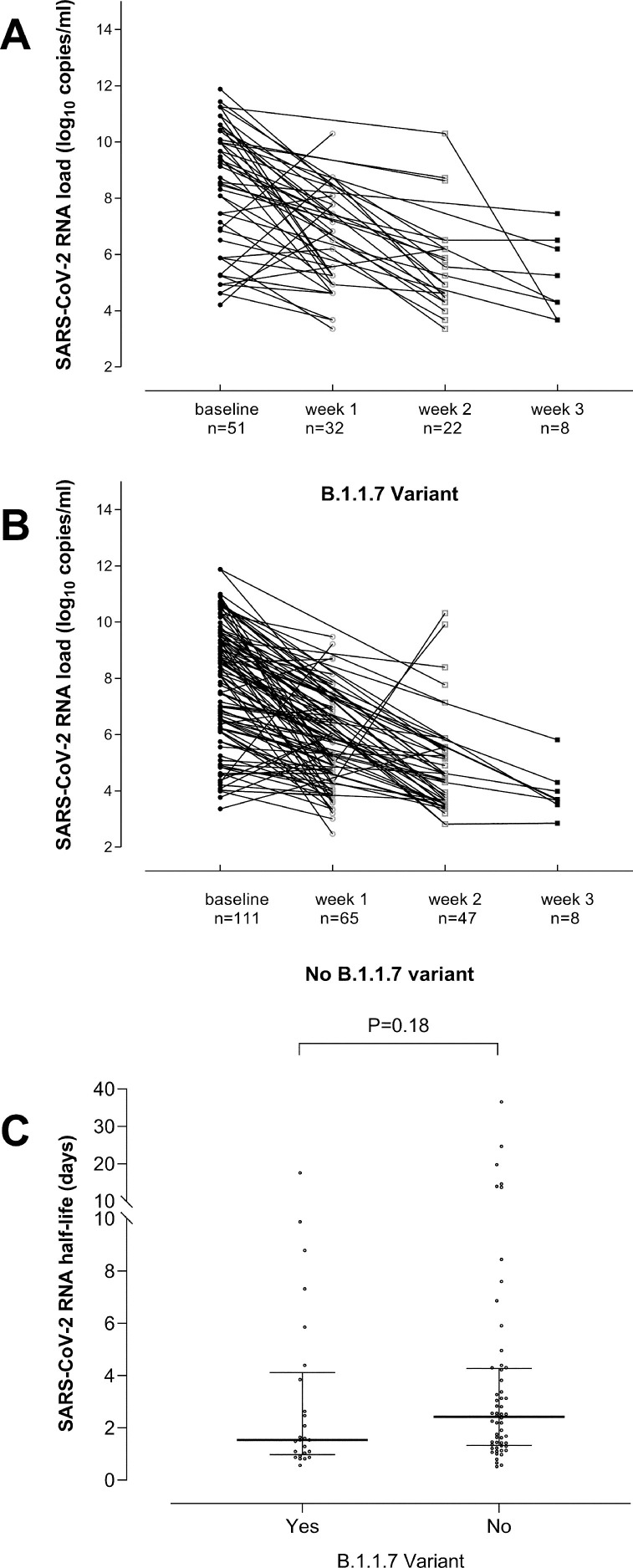
Fig. 2.
Kinetics of SARS-CoV-2 RNA load in nasopharyngeal specimens from individuals either infected by the B.1.1.7 variant (A) or by other variants (B) through week 3 after diagnosis of SARS-CoV-2 infection. Trajectories of SARS-CoV-2 RNA load in URT were classified as rising or decreasing, as the variation in the viral load between consecutive specimens was > or <0.5 log10 copies/ml (upper limit of intra-assay log10 variation), respectively. SARS-CoV-2 RNA half-life in the upper respiratory tract from individuals either infected by the B.1.1.7 variant or by other variants (C). The kinetics of SARS-CoV-2 RNA clearance followed a logarithmic decay curve in most individuals, expressed by the equation yt=y0e−kt, where y0 is the initial SARS-CoV-2 RNA load, t is time of follow-up specimen sampling since diagnosis (initial RT-PCR result) and k is the decay constant. SARS-CoV-2 RNA load half-life was then calculated using the equation ln2/k. SARS-CoV-2 RNA half-life in URT was calculated only for individuals (25 infected by the B.1.1.7 variant and 56 controls) that met two criteria: (i) displaying a descending trajectory throughout the study period (three weeks); (ii) the follow-up specimen used for calculations was collected within 4–10 days after the initial one. P value for comparisons across groups is shown.