Fig. 4. TDMD explains the instability of most short-lived miRNAs.
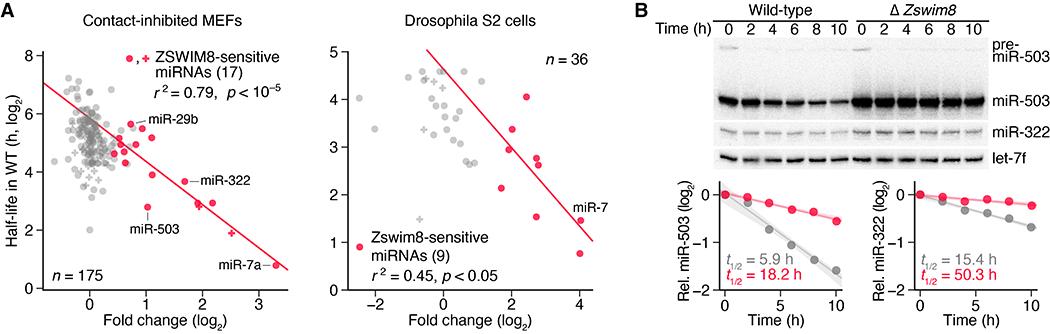
(A) The relationship between miRNA half-life and ZSWIM8 sensitivity. For each miRNA guide strand reliably quantified in contact-inhibited MEFs (left) or Drosophila S2 cells (right), its half-life in wild-type cells (4, 5) is plotted as function of the fold change in its mean level observed upon loss of ZSWIM8, as quantified in Fig. 3B. If both strands of a miRNA duplex accumulated to similar levels (within 5-fold of each other in Zswim8 knockout cells), implying that both strands commonly served as guide strands, then both strands were included in the analysis, with points indicated (+ instead of •). Points for ZSWIM8-sensitive miRNAs are colored red, as in Fig. 3B, and the line is the least-squares fit to these points (r2, coefficient of determination; p value indicated). (B) The influence of ZSWIM8 on miRNA decay. At the top is an RNA blot measuring miR-503, miR-322, and let-7f levels in NIH 3T3 polyclonal lines expressing Cas9 and either a non-targeting control gRNA (wild-type) or a Zswim8-targeting gRNA (table S1B). Serum was re-introduced to serum-starved cultures, and samples were taken at the indicated times. Plotted below are miR-503 and miR-322 levels in wild-type and Δ Zswim8 cells (gray and red, respectively), after normalization to the loading control (let-7f) and the 0 h timepoint of each cell line. Lines show the least-squares fit to the data (shading, 95% confidence interval), with apparent half-lives (t1/2 values) derived from the fit also indicated. n = 1 biological replicate.
