FIG 3.
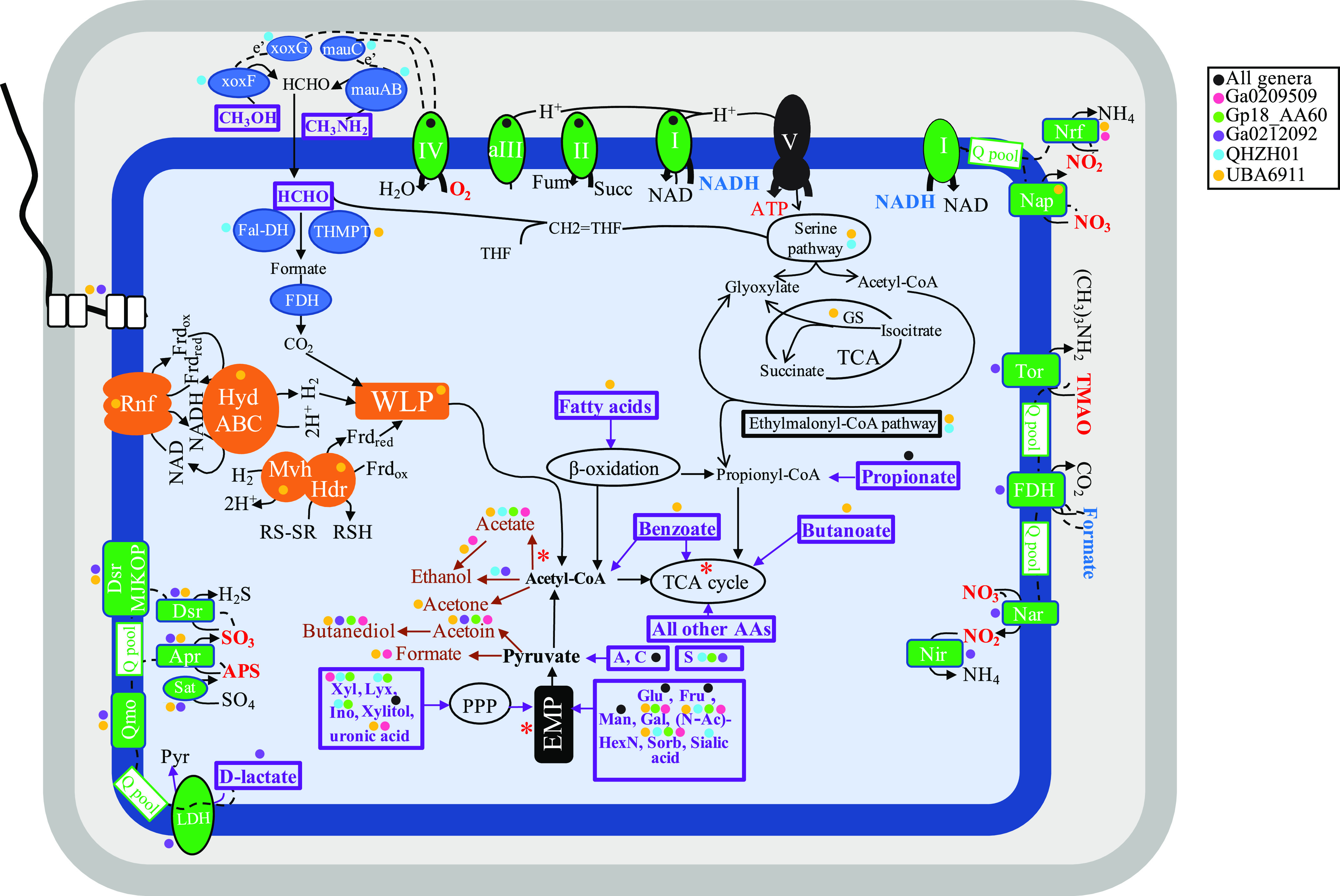
Cartoon depicting different metabolic capabilities encoded in family UBA6911 genomes with capabilities predicted for different genera shown as colored circles (all orders, black; genus Ga0209509, pink; genus Gp18_AA60, green; genus Ga0212092, purple; genus QHZH01, cyan; genus UBA6911, yellow). Enzymes for C1 metabolism are shown in blue. Electron transport components are shown in green, and electron transfer is shown as dotted black lines from electron donors (shown in boldface blue text) to terminal electron acceptors (shown in boldface red text). The sites of proton extrusion to the periplasm are shown as black arrows, as is the F-type ATP synthase (V). Proton motive force generation, as well as electron carrier recycling pathways associated with the operation of the WLP, is shown in orange. All substrates predicted to support growth are shown in boldface purple text within thick purple boxes. Fermentation end products are shown in brown. Sites of substrate-level phosphorylation are shown as red asterisks. A flagellum is depicted, the biosynthetic genes of which were identified in genomes belonging to the genera UBA6911 and Ga0212092. The cell is depicted as rod shaped based on the identification of the rod shape-determining proteins rodA, mreB, and mreC in all genomes. Abbreviations: Apr, the enzyme complex adenylylsulfate reductase (EC 1.8.99.2); APS, adenylyl sulfate; Dsr, dissimilatory sulfite reductase (EC 1.8.99.5); EMP, Embden-Meyerhoff-Parnas pathway; Fal-DH, formaldehyde dehydrogenase; FDH, formate dehydrogenase; Frdox/red, ferredoxin (oxidized/reduced); Fru, fructose; fum, fumarate; Gal, galactose; Glu, glucose; GS, glyoxylate shunt; Hdr, heterodisulfide reductase complex; HydABC, cytoplasmic [Fe Fe] hydrogenase; I, II, aIII, and IV, aerobic respiratory chain comprising complex I, complex II, alternate complex III, and complex IV; Ino, myo-inositol; LDH, l-lactate dehydrogenase; Lyx, lyxose; Man, mannose; mauABC; methylamine dehydrogenase; Mvh, Cytoplasmic [Ni Fe] hydrogenase; Nap, nitrate reductase (cytochrome); Nar, nitrate reductase; Nir, nitrite reductase (NADH); Nrf, nitrite reductase (cytochrome c-552); N-AcHexN, N-acetylhexosamines; PPP, pentose phosphate pathway; Pyr, pyruvate; Q pool, quinone pool; Qmo, quinone-interacting membrane-bound oxidoreductase complex; RNF, membrane-bound RNF complex; RSH/RS-SR, reduced/oxidized disulfide; Sorb, sorbitol; succ, succinate; TCA, tricarboxylic acid cycle; THMPT, tetrahydromethanopterin-linked formaldehyde dehydrogenase; TMAO, trimethylamine N-oxide; Tor, trimethylamine-N-oxide reductase (cytochrome c); V, ATP synthase complex; WLP, Wood-Ljungdahl pathway; xoxFG, methanol dehydrogenase; Xyl, xylose.
