Figure 6. Residual cBAF complexes secondary to ARID1A/B loss do not possess neo-functionality.
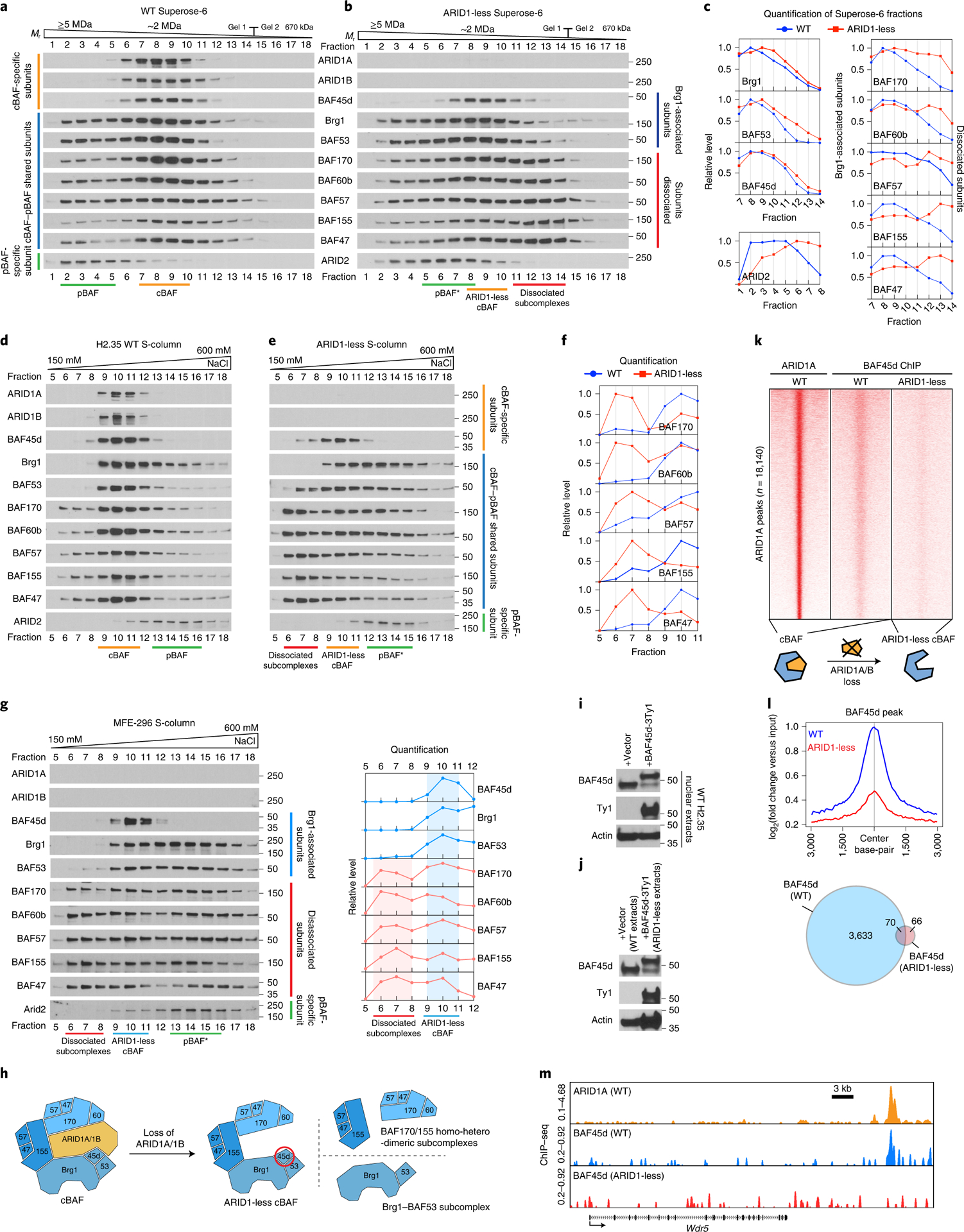
a. Gel filtration chromatography of control H2.35 cell nuclear extracts using a Superose-6 column.
b. Gel filtration chromatography of ARID1-less H2.35 cell nuclear extracts.
c. Quantification of western blot data in a and b.
d. Cation exchange chromatography of control H2.35 nuclear extracts using an S-column.
e. Cation exchange chromatography of ARID1-less H2.35 nuclear extracts.
f. Quantification of western blot data in d and e.
g. Cation exchange chromatography of MFE-296 nuclear extracts and the corresponding quantification.
h. Model for three types of subcomplexes resulting from cBAF dissociation in the absence of ARID1 paralogs. The ARID1-less cBAF specific subunit BAF45d is circled in red.
i. Expression of Ty1 tagged BAF45d in control H2.35 cells.
j. Expression of Ty1 tagged BAF45d in ARID1-less H2.35 cells. The expression level was compared with endogenous BAF45d in WT extracts due to partial loss of endogenous BAF45d in ARID1-less cells.
k. The occupancies of BAF45d in control and ARID1-less cells, which represent control cBAF and ARID1-less cBAF complex binding, shown as a heatmap. ARID1A binding loci were used as a reference.
l. The corresponding averaged ChIP-seq peaks and Venn diagram are shown.
m. Representative genome browser tracks showing impaired binding of ARID1-less cBAF (BAF45d peaks in ARID1-less cells).
