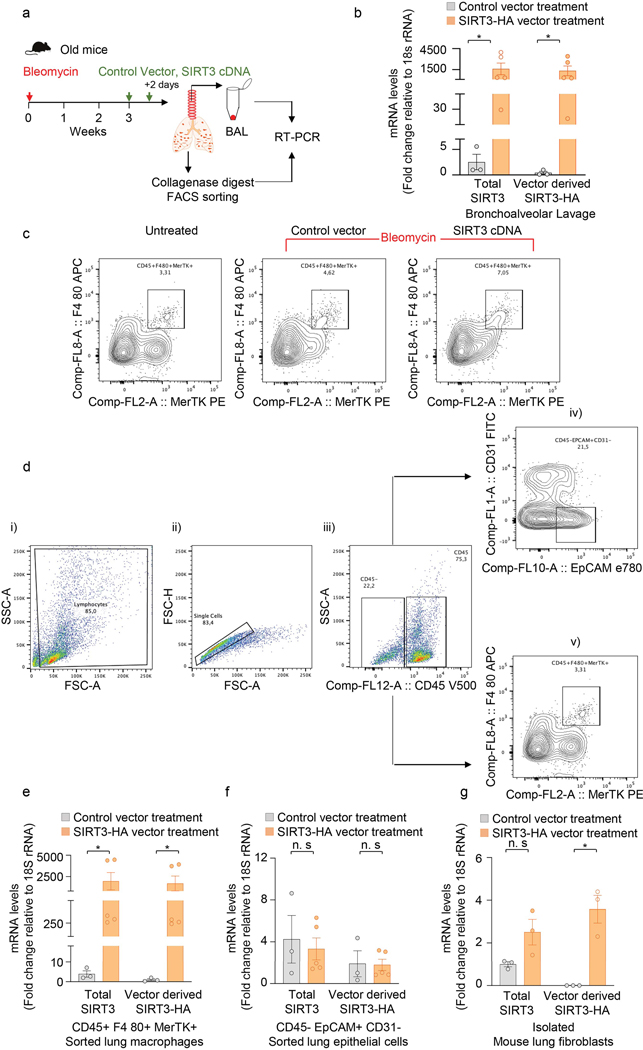
Extended Data Fig. 2. Exogenous SIRT3 cDNA is preferentially expressed in lung macrophages.
(a) Schematic of experiment design. (b) RT-PCR analysis of total SIRT3 or vector derived SIRT3-HA mRNA levels in bronchoalveolar lavage (BAL) from mice subjected to bleomycin injury followed by treatment with control vector (n = 3) or SIRT3 cDNA plasmid (n = 5), as indicated in the Methods section. Data presented as means ± s.e.m., *P =0.0357 (unpaired t-test, non-parametric, two-tailed). (c) Flow cytometric analysis of macrophages (CD45+ F4 80+ MerTK+) sorted from collagenase lung digests of naïve mice and mice injured with bleomycin followed by treatment with control vector or SIRT3 cDNA are shown. (d) Gating strategy: (i) cells were gated for FSC-A against SSC-A; (ii) doublets were excluded using FSC-H against FSC-A; (iii) singlets were gated for CD45 positive/negative population; (iv) within the CD45- population, epithelial cells were gated as EPCAM+CD31-; and (v) within the CD45+ population, macrophages were gated as F480+MerTK+. (e-g) RT-PCR analysis of total SIRT3 or vector derived SIRT3-HA mRNA levels in FACS sorted macrophages (e) and epithelial cells (f), and adherence-purified fibroblasts (g) from whole lung collagenase digest from mice subjected to bleomycin injury followed by treatment with control vector (n = 3) or SIRT3 cDNA plasmid (n = 5). Data presented as means ± s.e.m., *P = 0.0357 (macrophages;unpaired t-test, non-parametric, two-tailed), P = 0.0052 (fibroblasts; unpaired t-test, two-tailed statistical analysis), n.s. = not significant.