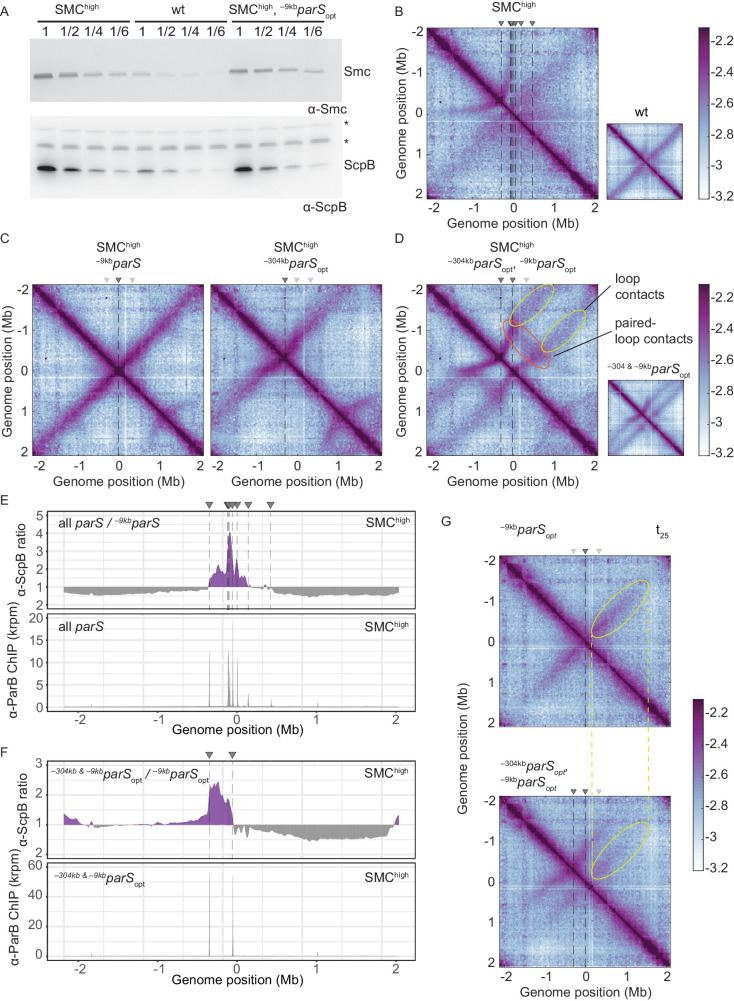
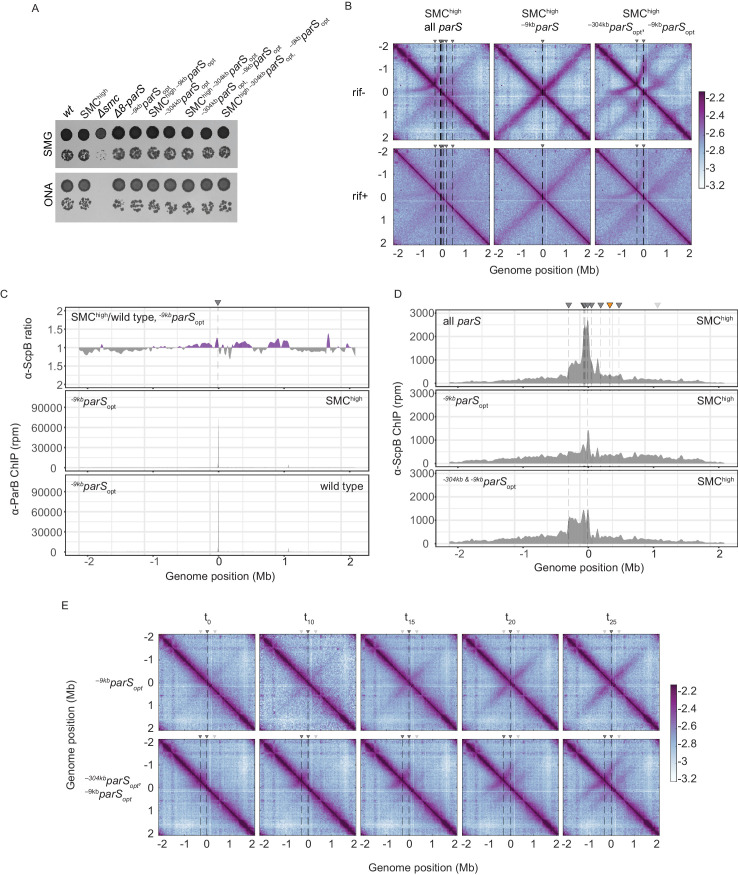
Figure 4. Increasing the cellular pool of Smc hampers chromosome organization.
(A) Immunoblotting using α-Smc (top panel) and α-ScpB serum (bottom panel). SMChigh denotes strains with extra genes for Smc-ScpAB. Protein extracts of wild-type or SMChigh strains (harboring all parS sites or single parS site) were serially diluted with extracts from Δsmc or ΔscpB strains as indicated (see Materials and methods). * indicates unspecific bands generated by the α-ScpB serum. (B) Normalized 3C-seq contact map for SMChigh strain with all parS sites present. Inset shows 3C-seq contact map of a strain with wild-type protein levels (also displayed in Figure 1C) for direct comparison. (C) Normalized 3C-seq contact maps for SMChigh strains with parSopt at −9 kb only or at −304 kb only. (D) Normalized 3C-seq contact map for SMChigh strain with parSopt at positions: −9 kb and −304 kb. As in (B), with inset displaying respective control strain with normal Smc expression levels (also shown in Figure 3B). (E) Ratio plots for ChIP-seq read counts comparing SMChigh strains with all parS sites and a single parS site (–9kbparSopt). Representation as in Figure 2B (top panel). Read count for α-ParB ChIP-seq in SMChigh strain (bottom panel). (F) As in (E) involving a SMChigh strain with two parS sites (–304kbparSopt and –9kbparSopt) instead of all parS sites. (G) Normalized 3C-seq contact maps for time point t25 after IPTG-induced ParB expression with a single parSopt site (top panel) or two parSopt sites (at −9 kb and −304 kb) (bottom panel). Ellipsoids (in yellow colors) mark the position of contacts stemming from loop extrusion originating at –9kbparSopt.