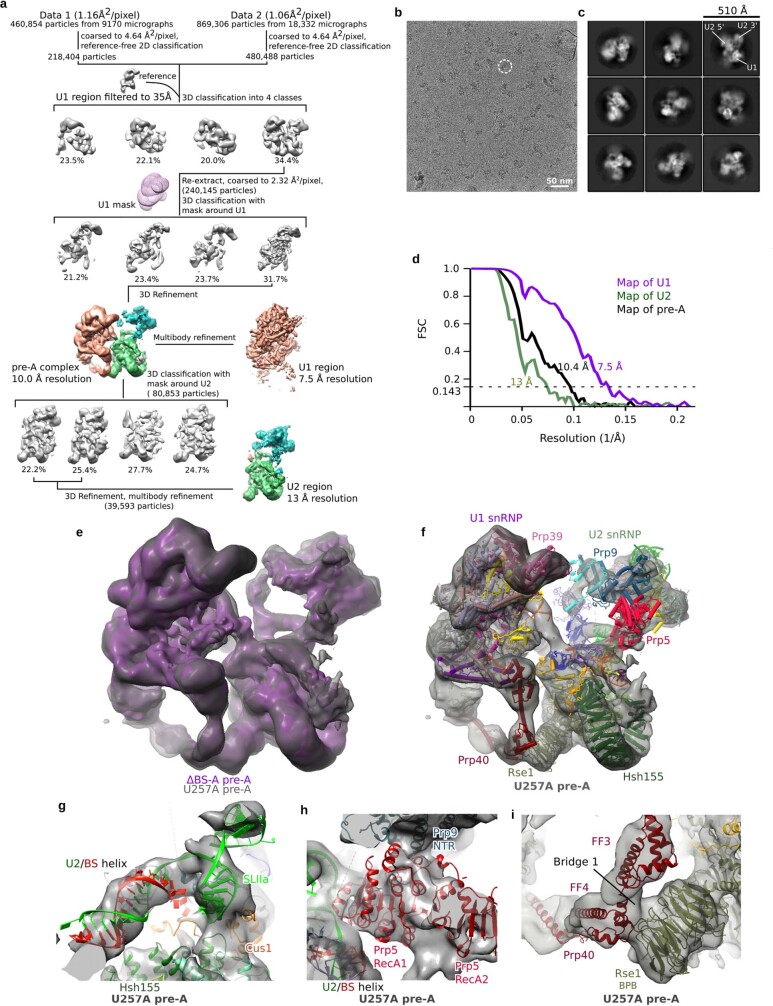
Extended Data Fig. 7. Cryo-EM and image-processing of the U257A pre-A complex.
a, Computation sorting scheme, with all major image-processing steps depicted. For a more detailed explanation, see the Methods section on ‘Image processing’. b, Typical cryo-EM micrograph (out of a total of 27,502) of the S. cerevisiae U257A pre-A complex recorded at ×120,700 magnification with a Titan Krios microscope using a Falcon III direct electron detector operating in integration mode at a calibrated pixel size of 1.16 Å. c, Representative cryo-EM 2D class averages of the yeast U257A pre-A complex. d, FSC calculated using the ‘Post-processing’ routine in RELION indicates a global resolution of 10.4 Å for the entire yeast U257A pre-A complex, and resolutions of 7.5 Å and 13 Å for the multibody-refined U1 and U2 regions, respectively. The global resolution was lower than that of the ΔBS-A pre-A complex, mainly because of the lower number of particles analysed. e, Overlay of the EM densities of the ΔBS-A (purple) and U257A (grey) pre-A complexes. f, Fit of the 3D model of the ΔBS-A pre-A complex into the EM density of the U257A pre-A complex. Note that, for both complexes, density encompassing Prp5 is first observed at a lower threshold. An extended U2–BS helix has also formed in complexes formed on the U257A mutant. However, the precise conformation of the helix cannot be discerned. The Hsh155HEAT domain is in an open conformation and Prp5 is still bound at the same position, and the same U1–U2 bridges are also observed, indicating that the U257A complexes are also stalled at the same pre-A stage. g, Fit of the extended U2–BS helix from the ΔBS-A pre-A complex into the U257A pre-A EM density. h, Fit of the Prp5RecA domains and U2–BS helix from the ΔBS-A pre-A model into the EM density of the U257A pre-A complex. i, Fit of the Prp40 FF domains and Rse1BPB (which comprise bridge 1) from the ΔBS-A pre-A model into the EM density of the U257A pre-A complex.