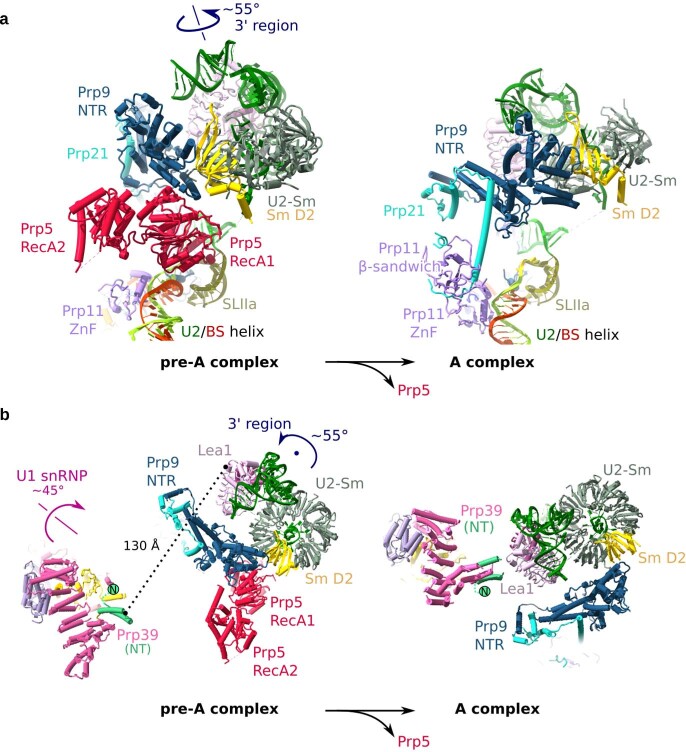
Extended Data Fig. 8. Movement of U1 and U2 snRNPs during the transition from the pre-A to the A complex.
a, Close-up of the rotation of the U2 3′-region after the release of Prp5. The 3′-region rotates around the indicated axis by roughly 55°. To better show the movement of the 3′-region, the SmD2 protein is in yellow. For simplicity, only the 3′-region of U2 plus U2 SLII and the U2–BS helix are shown in the pre-A complex and the yeast A complex (PDB 6G90). The pre-A and A complexes are aligned via U2 SLIIa and HR 19–20 of Hsh155. b, Close-up of the movement of the U1 snRNP and 3′-region of U2. A top view is shown, with the black dot indicating the pivot point of the U2 3′-region, which rotates by roughly 55° in the plane of the paper. For simplicity, only the region of U1 snRNP that contains Prp39 is shown. The U1 snRNP rotates around the indicated axis by roughly 45°. In the pre-A complex, Prp39 and Lea1 are separated by roughly 130 Å, but the movements of U1 and U2 bring them into close proximity in the A complex. Even though Lea1 is not essential in S. cerevisiae, its depletion prevents formation of the A complex, and adding back Lea1 restores A-complex assembly47. The Prp39–Lea1 interaction is a structural marker for the formation of a mature A complex, and as such its absence in the pre-A complex is a clear indication that our complex has stalled at an earlier assembly stage. This interaction is also maintained in the pre-B complex13 and is therefore also a structural marker for the conformation that allows joining of the tri-snRNP.