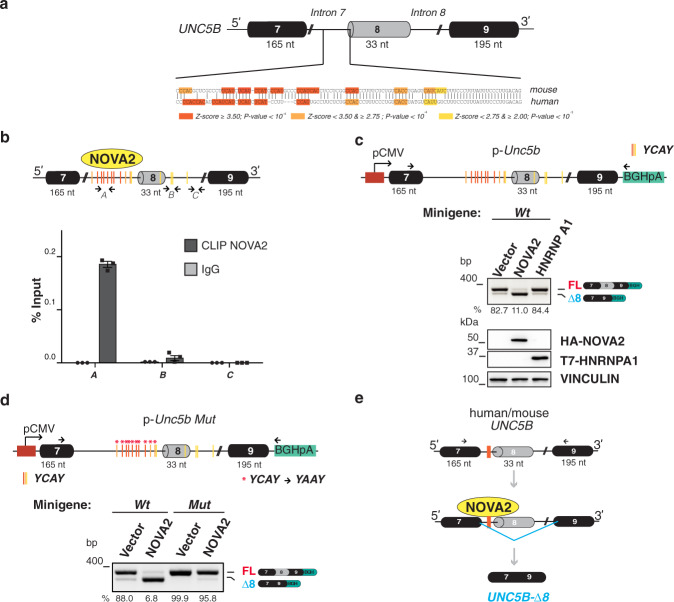
Fig. 2. NOVA2 directly binds to Unc5b pre-mRNA.
a Schematic representation of mouse and human UNC5B pre-mRNAs. AS exon 8 in gray. Constitutive exons 7 and 9 in black. Introns 7 and 8 are also indicated (thin lines). YCAY sites, identified by using the RBPmap tool within 200 nt from the exon–intron junctions, are indicated in orange, pale orange, or yellow bars depending on their Z-score and P values (calculated by RBPmap tool). b Upper: schematic representation of mouse Unc5b as in a. Arrows indicate primers used for RT-qPCR of NOVA2 immunoprecipitated RNAs. Lower: RT-qPCR of NOVA2 CLIP experiments in moEC. Amplification of immunoprecipitated RNAs with primers annealing to the YCAY cluster in intron 7 (A) and, as negative controls, within intron 8 (B and C). Error bars indicate ±SD calculated from n = 3 independent experiments. c Upper: the mouse Unc5b genomic region encompassing exons 7, 8, and 9, cloned into the pcDNA3.1(+) vector to generate the p-Unc5b wild-type (Wt) minigene. Arrows indicate primers used for splicing analysis of the transcripts generated from the minigene. Boxes = exons; thin lines = introns; pCMV = promoter; BGHpA = polyadenylation sequence. Colored vertical bars as in a. Lower: p-Unc5b Wt co-transfected in moEC with either NOVA2-HA (NOVA2), T7-HNRNPA1 (HNRNPA1), or empty vector (Vector). RT-PCR analysis of p-Unc5b splicing is shown. Ectopic expression of NOVA2 and HNRNPA1 were confirmed by immunoblotting with anti-HA and anti-T7 antibodies, respectively. VINCULIN as loading control. d Splicing assay with p-Unc5b Mut in moEC. Red asterisks indicate the mutated NOVA2 binding sites (YCAY repeats were replaced with YAAY). e NOVA2 (yellow circle) promotes UNC5B exon 8 skipping by directly binding to the YCAY cluster (orange bar) located in the intronic region upstream exon 8. The percentage of exon inclusion is shown below each gel. At least three independent biological replicates were analyzed for each experiment.