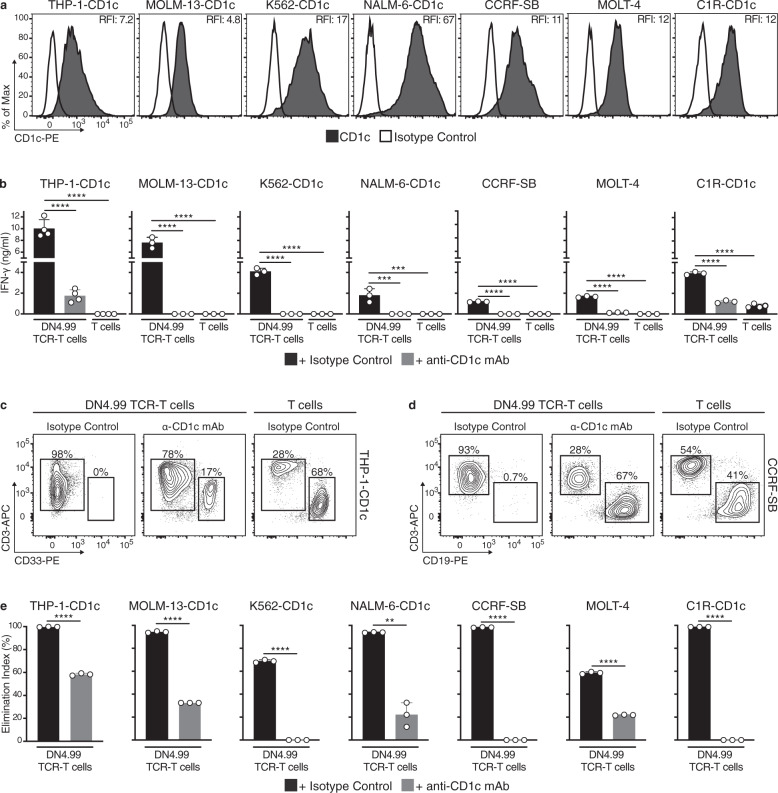
Fig. 4. DN4.99 TCR-T cells efficiently recognize and kill CD1c+ leukemia-cell lines.
Primary T cells were transduced (DN4.99 TCR-T cells) or not-transduced (T cells) with the lead DN4.99 TCR and assessed in vitro for the recognition and killing of CD1c-expressing acute leukemia or lymphoblastoid cell lines. a CD1c expression (black histograms) on 7 different acute leukemia (THP-1, MOLM-13, K562, NALM-6, CCRF-SB, MOLT-4) or lymphoblastoid (C1R) cell lines, as determined by flow cytometry measurement of labeling with anti-human CD1c monoclonal antibody (mAb). White histograms represent labeling with isotype-matched control mAb. Relative Fluorescence Intensity (RFI) was calculated as the ratio between intensity of labeling of the sample and control. b Recognition of CD1c-expressing acute leukemia or lymphoblastoid cell lines by DN4.99 TCR-transduced or non-transduced primary T cells following 48 h of co-culture at a 1:1 Effector:Target ratio with either 20 μg/ml of blocking anti-CD1c mAb (gray bars) or an equivalent concentration of mouse IgG1κ isotype control (black bars). IFN-γ secretion into supernatants was measured by ELISA. Data are represented as mean ± SD. Results are representative of 2 (for CCRF-SB, MOLT-4 and C1R-CD1c) and 4 (for THP-1-CD1c, MOLM-13-CD1c, K562-CD1c and NALM-6-CD1c) independent experiments with n = 3 replicates per experiments. ***P = 0.0001; **** P < 0.0001 determined by Ordinary one-way ANOVA followed by Tukey’s multiple comparison test. c, d Representative flow cytometry analysis to assess killing of THP-1-CD1c (c) and CCRF-SB (d) cells by DN4.99 TCR-T cells after 72 h of the same co-culture conditions of (b). e Killing of each target cell line in the same co-cultures determined as shown in (c, d), expressed as an Elimination Index as described in Methods. Gray bars represent co-cultures with anti-CD1c mAb, and black bars with the isotype control. Data are represented as mean ± SD. Results are representative of 2 (for K562-CD1c, CCRF-SB, MOLT-4 and C1R-CD1c) and 4 (for THP-1-CD1c, MOLM-13-CD1c and NALM-6-CD1c) independent experiments with n = 3 replicates per experiments. **P = 0.0071; ****P < 0.0001 determined by unpaired two-tailed Welch’s t-test.