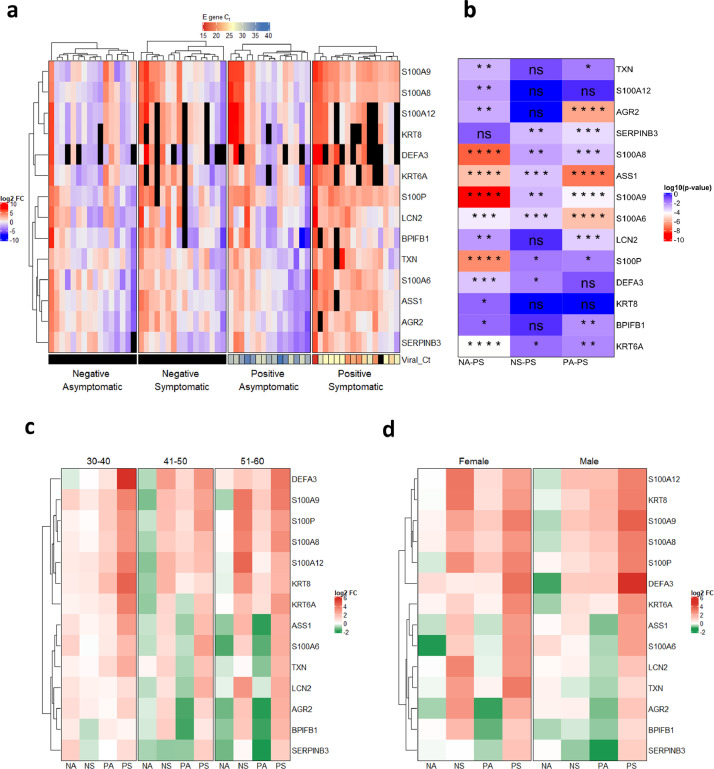
Fig. 3.
qRT-PCR validated expression profile of selected genes in different categories of COVID-19 cohort. a) qRT-PCR was performed on RNA isolated from COVID-19 patients for 14 genes and average log2FC values (with respect to Negative Asymptomatic group) of PCR triplicates are shown in a heatmap. Each column represents a patient. The bottom annotation shows the Ct value for the viral gene encoding Envelope (E) protein with a corresponding legend on the top. Black boxes denote 'value unknown/undetermined'. b) Differences between groups for each gene were computed and the log10 (p-value) of comparisons is shown in the heatmap. The comparisons are Negative asymptomatic vs Positive symptomatic (NA-PS), Negative symptomatic vs Positive symptomatic (NS-PS), and Positive asymptomatic vs Positive symptomatic (PA-PS). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; ns – not significant (Kruskal-Wallis test followed by post hoc Dunn's test with Bonferroni corrections for multiple comparisons). c) log2FC values are grouped based on age groups 30-40, 41-50, and 51-60. Each row represents the average of log2FC values for patients falling into the particular age group and respective disease status. d) log2FC values are grouped according to sex. Each row represents the average of log2FC values for patients falling into the particular sex and respective disease status.