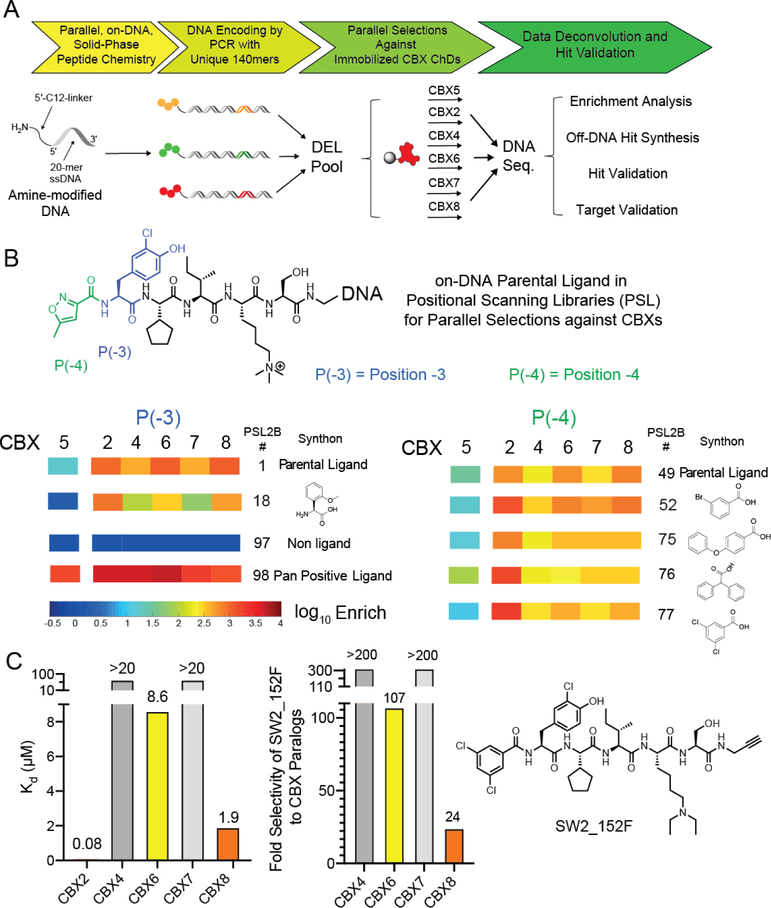
Figure 3:
A) Preparation and selection of DNA-encoded chemical libraries for CBX ChD ligands. Positional scanning libraries (PSL) were prepared by parallel chemical modification of an amine-modified oligonucleotide immobilized on a solid support. Subsequent encoding was performed by parallel PCRs with unique templates. Libraries were pooled and selected against immobilized CBX ChDs. Enriched libraries were then additionally barcoded with index sequences, pooled, and sequenced. B) Representative monomers with increased affinity or selectivity are depicted and enrichment heat map from previous selections were illustrated. Enrichment was normalized to a non-ligand control. C). In vitro affinity determination of optimal CBX2 ligand SW2_152F using fluorescence polarization assay with fluorescein-conjugated ligand. Selectivity was calculated from Kd values (Table 1).