FIGURE 1.
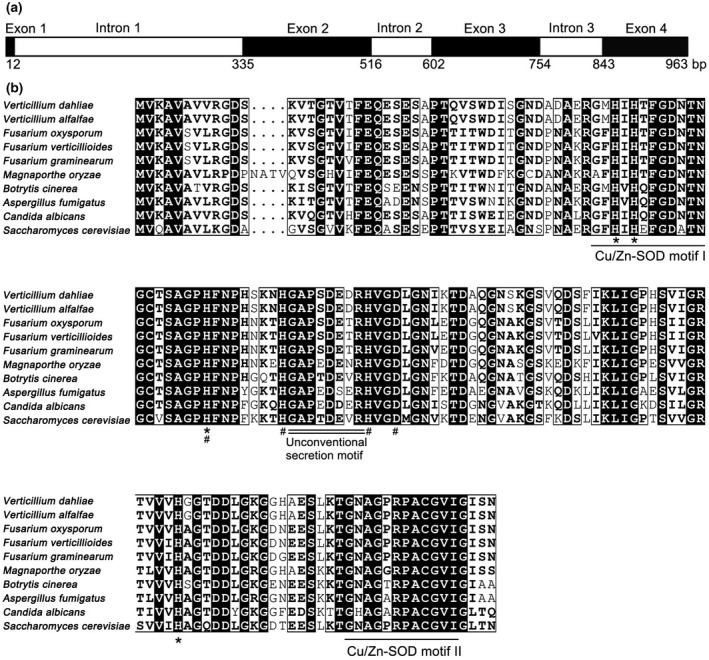
Bioinformatic analysis of VdSOD1. (a) Structure of VdSOD1. Exons are black boxes and introns are white boxes. (b) Alignment of VdSOD1 and homologs from other fungi. Highly conserved residues are marked by black boxes. The Cu/Zn‐SOD motifs are underlined, Cu2+ and Zn2+ binding sites are marked by asterisks and hashtags, respectively. The unconventional secretion motif is double‐underlined. GenBank accession number of aligned sequences are Verticillium dahliae (VEDA_03436), Verticillium alfalfae (XP_003007974.1), Fusarium oxysporum (EGU80493.1), Fusarium verticillioides (EWG38836.1), Fusarium graminearum (CEF77131.1), Magnaporthe oryzae (XP_003721165.1), Botrytis cinerea (XP_001560530.1), Aspergillus fumigatus (Q9Y8D9.3), Candida albicans (EEQ45246.1), and Saccharomyces cerevisiae (NP_012638.1)
