FIGURE 8.
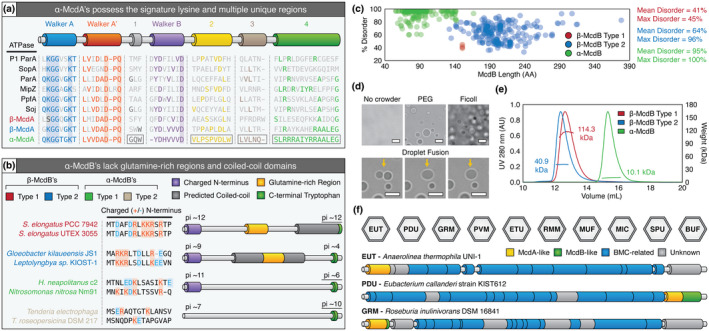
Similarities and differences among all known McdA and McdB proteins. (a) Features conserved or distinct among α‐McdA (green), β‐McdA Type 1 (red), β‐ McdA Type 2 (blue), and diverse classical ParA‐type proteins. All Walker‐boxes are well‐conserved among all classic ParA family proteins. ParA‐type proteins shown: Escherichia coli phage P1 ParA (plasmid partitioning—YP_006528) (Abeles et al., 1985), Escherichia coli F plasmid SopA (plasmid partitioning—NP_061425) (Mori et al., 1986), Caulobacter crescentus ParA (chromosome segregation—AAB51267) (Mohl & Gober, 1997), Caulobacter crescentus MipZ (cell‐division positioning—NP_420968) (Thanbichler & Shapiro, 2006), Rhodobacter sphaeroides PpfA (chemotaxis cluster distribution—EGJ21499) (Roberts et al., 2012), and Bacillus subtilis Soj (chromosome segregation—NP_391977) (Marston & Errington, 1999). (b) α‐McdB proteins lack the predicted central coiled‐coil and glutamine‐rich regions found in β‐McdB proteins. α‐McdB Type 2 proteins lack the charged N‐terminus conserved in all other McdB types. (c) PONDR disorder scatter plot for all McdB protein types. (d) DIC microscopy images showing purified H. neapolitanus McdB undergoes LLPS in vitro in the presence of the crowders PEG or Ficoll. Droplets exhibit liquid‐like properties such as fusion (yellow arrows). (e) SEC‐MALS plot for a representative β‐McdB Type 1 (S. elongatus McdB; monomer MW = 17 kDa), β‐McdB Type 2 (Synechococcus sp. PCC 7002 McdB; monomer MW = 21 kDa), and α‐McdB (H. neapolitanus McdB; monomer MW = 10 kDa). (f) McdA/B‐like sequences genomically neighbor BMC components across diverse microbes [Colour figure can be viewed at wileyonlinelibrary.com]
