FIGURE 1.
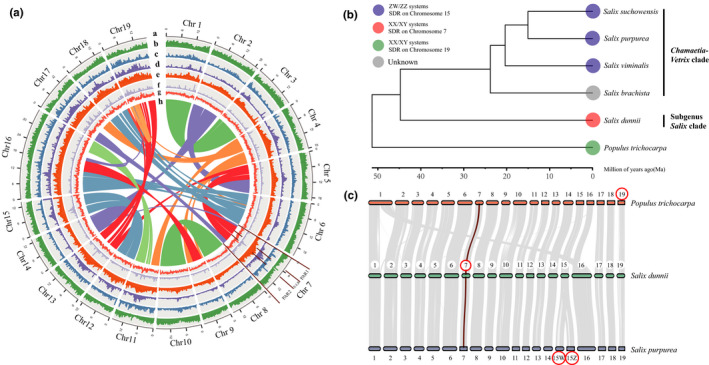
Genome structure and evolution of Salix dunnii. a, circos plot showing: (a) the chromosome lengths in Mb, (b) gene density, (c) LTR‐Copia density, (d) LTR‐Gypsy density, (e) total repeats, (f) density of pseudogenes, (g) GC (guanine‐cytosine) content and (h) syntenic blocks. b, Inferred phylogenetic tree of S. brachista, S. dunnii, S. purpurea, S. suchowensis, S. viminalis and the outgroup Populus trichocarpa, with divergence times. The root age of the tree was calibrated to 48–52 million years ago (Ma) following Chen et al. (2019) and the crown age of the Chamaetia‐Vetrix clade (here including S. brachista, S. purpurea, S. suchowensis, and S. viminalis) was calibrated to 23–25 Ma according to Wu et al. (2015). c, Macrosynteny between genomic regions of P. trichocarpa, S. dunnii and S. purpurea. The dark orange line shows the syntenic regions between the S. dunnii X‐linked region of chromosome 7, and the homologous regions in the same chromosomes of S. purpurea and P. trichocarpa. Red circles show the chromosomes carrying sex‐linked regions
