FIGURE 3.
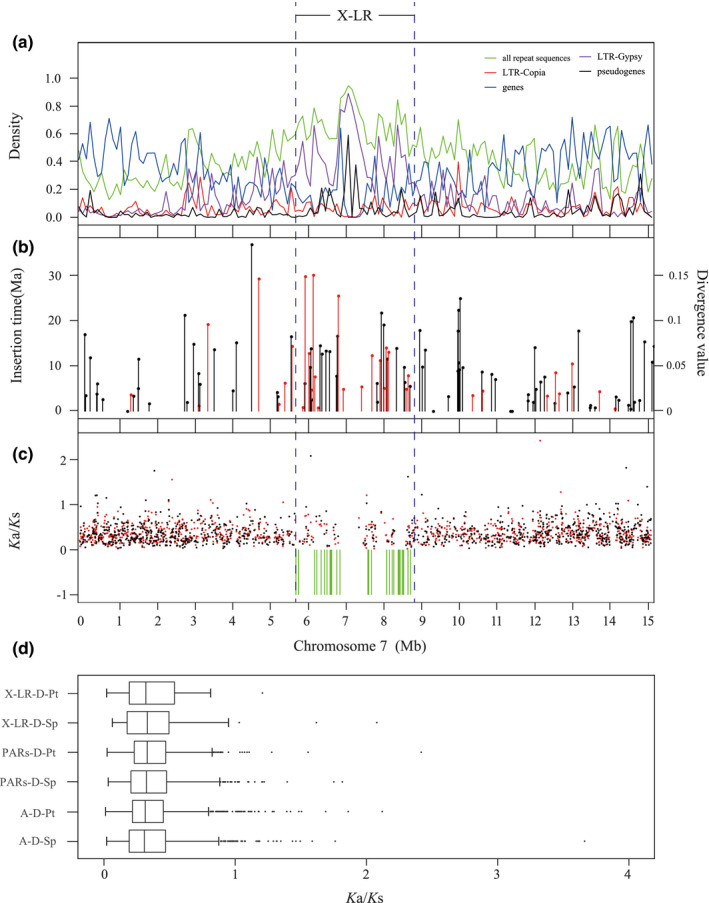
Analysis of Salix dunnii chromosome 7 genes. (a) Densities of two transposable element types, LTR‐Gypsy (purple line) and LTR‐Copia (red line), all repeat sequences (green line), pseudogenes (black line), as well as genes (blue line) in the entire chromosome 7 of S. dunnii. (b) Estimated insertion times and divergence values of full‐length long terminal repeat retrotransposons (LTR‐RTs) in chromosome 7 of S. dunnii. The red lines represent LTR‐Gypsy, and the black lines LTR‐Copia elements. (c) Comparison of Ka/Ks ratios between homologous genes in S. dunnii and Populus trichocarpa (red dots), and of S. dunnii vs. S. purpurea (black dots). Green lines indicate locations of S. dunnii X‐linked genes with no hits in either S. purpurea or P. trichocarpa. (d) Comparison of Ka/Ks values of X‐LR, PARs and autosomal genes (chromosome 6). X‐LR‐D‐Pt and PARs‐D‐Pt are obtained from the homologous genes of S. dunnii and P. trichocarpa. X‐LR‐D‐Sp and PARs‐D‐Sp are obtained from chromosome 7 of the homologous genes of chromosome 7 of S. dunnii and S. purpurea. A‐D‐Pt and A‐D‐Sp are obtained from the homologous genes of chromosome 6 of S. dunnii – P. trichocarpa (1897 homologous pairs) and S. dunnii – S. purpurea (1852 homologous pairs), respectively. The Wilcoxon rank sum test was used to detect significance differences of different regions of the two data sets. No significant difference (p < .05) was detected between the sex‐linked region and the autosomes or PARs (Figure S19)
