Figure 7.
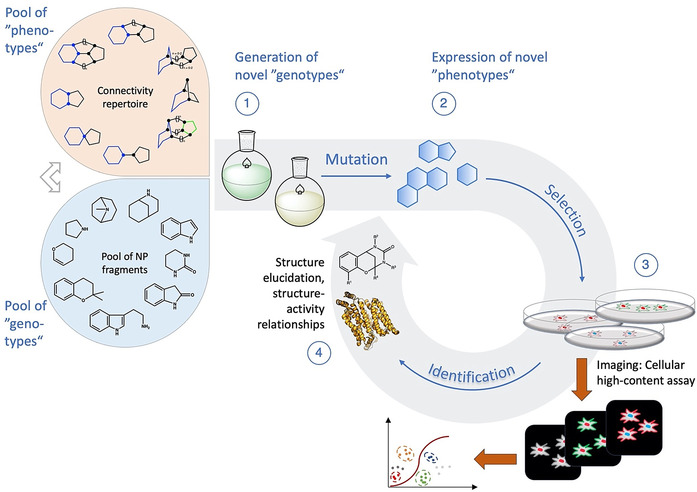
Focused chemical evolution. (1) The cyclic procedure starts with the generation of a “pool of genotypes” which is synthesised from NP fragments (=inheritable chemical information) using synthetic strategies that allow for the combinatorial application of a set of feasible connectivity patterns as well as further derivatisations. (2) Structural properties of the resulting compound library, such as content of sp3‐hybridised atoms, stereocentres, heteroatoms, and aromaticity, account for the expression of “phenotypes”—the potential to interact with specific structural motifs of proteins. (3) The compound library then is applied to a cellular screening platform (e.g., yeast cells) which is optically monitored for structural changes, e.g., by fluorescence imaging. Data are combined, analysed, and sorted according to selective constraints. (4) Molecules causing a desired change in the cellular system are isolated and identified (NMR, MS) and submitted to further structural characterisation, for example, by co‐crystallisation with putative target proteins (example used here: 4PYP[67]). The outcome of this round is the beginning of a new cycle which includes the “new chemical information” that was received.
