Figure 4.
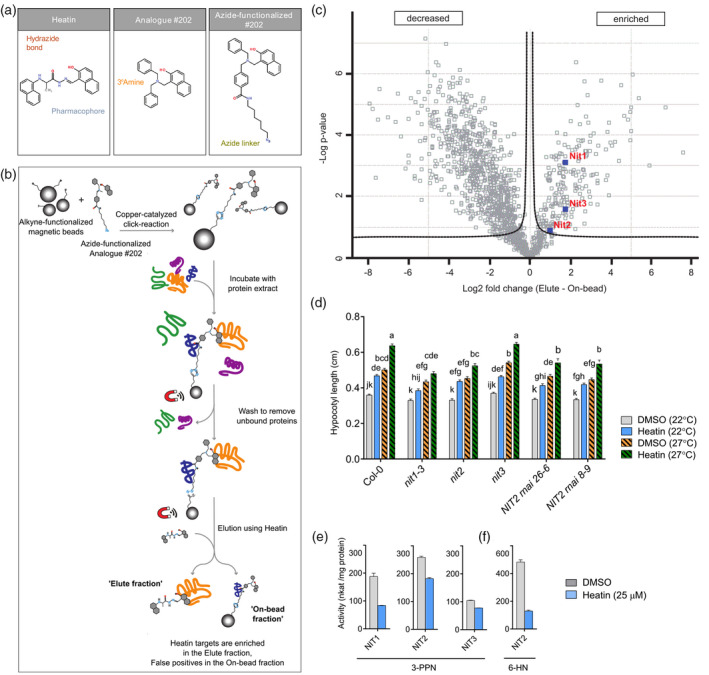
Nitrilase 1‐subfamily members are direct Heatin targets.
(a) Chemical structures of Heatin (left), analogue no. 202 (middle) and azide‐functionalized compound no. 202 (right). Highlighted are the pharmacophore (blue), Heatin’s hydrazide bond (red), analogue no. 202’s corresponding amine bond (yellow) and the azide linker (green).
(b) Schematic representation of the chemical proteomics strategy.
(c) Volcano plot of statistical significance against fold‐change of protein group label‐free quantification intensities between ‘Heatin‐eluted’ and ‘On‐bead fractions’ based on a two‐sided Student’s t‐test (FDR: 0.05; S0: 0.1). NIT1‐subfamily member proteins (indicated in blue squares and red letters) are enriched in the Elute fraction. Dotted lines represent the threshold for significant differences in protein abundances. Data are based on four biological replicates per condition.
(d) Hypocotyl lengths of 8‐day‐old nitrilase1‐subfamily mutant seedlings and Col‐0 wild type, grown on mock [dimethyl sulphoxide (DMSO), grey and orange bars] or in presence of Heatin (8.5 µm; blue and green bars), at either 22°C (open bars) or 27°C (dashed bars). Values are averages of six independent repetitions of 20–30 seedlings each. Letters indicate significance groups (Tukey HSD post‐hoc test), where averages that do not share letters are significantly different from each other (P < 0.05).
(e,f) In vitro enzymatic activity of recombinant NIT1‐subfamily proteins with (e) 3‐phenylpropionitrile (3‐PPN) or (f) 6‐heptenenitrile (6‐HN) as substrate (2.5 µm), with DMSO solvent as mock (grey bars) or Heatin (25 µm; blue bars), present in the reaction mix. Values are averages of three technical replicates. Error bars indicate SEM.
