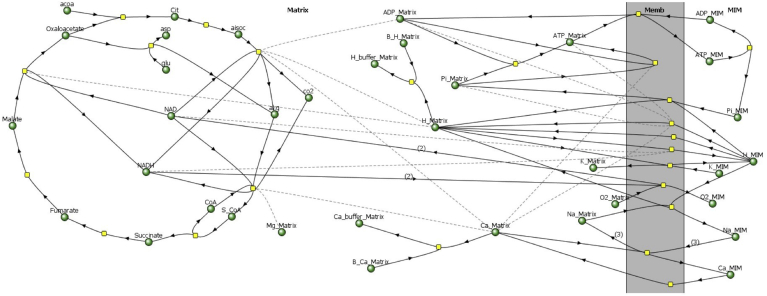
Fig. 1.
Diagram of the complete BioModel for mitochondrial energy metabolism used for simulations in Virtual Cell. Reaction pathways (solid arrows) are represented in terms of chemical species (metabolites, ions and buffers; green circles), and enzymes or transporters (yellow squares), based on the model of Nguyen et al. (2007). Effectors are connected to enzymes by dashed lines. The reactions occur in two compartments, the Matrix (left) and the space outside the matrix (right, MIM), and on the mitochondrial inner membrane (Memb) that separates them. Other non-standard abbreviations: acoa, acetyl Coenzyme A; aisoc, iso-citrate; akg, α-ketoglutarate; asp, aspartate; B_Ca, Ca2+ bound to calcium buffer (Ca_buffer); B_H, H+ bound to proton buffer (H_buffer); Cit, citrate; co2, carbon dioxide; glu, glutamate; O2, dioxygen; S_CoA, succinyl Coenzyme A.