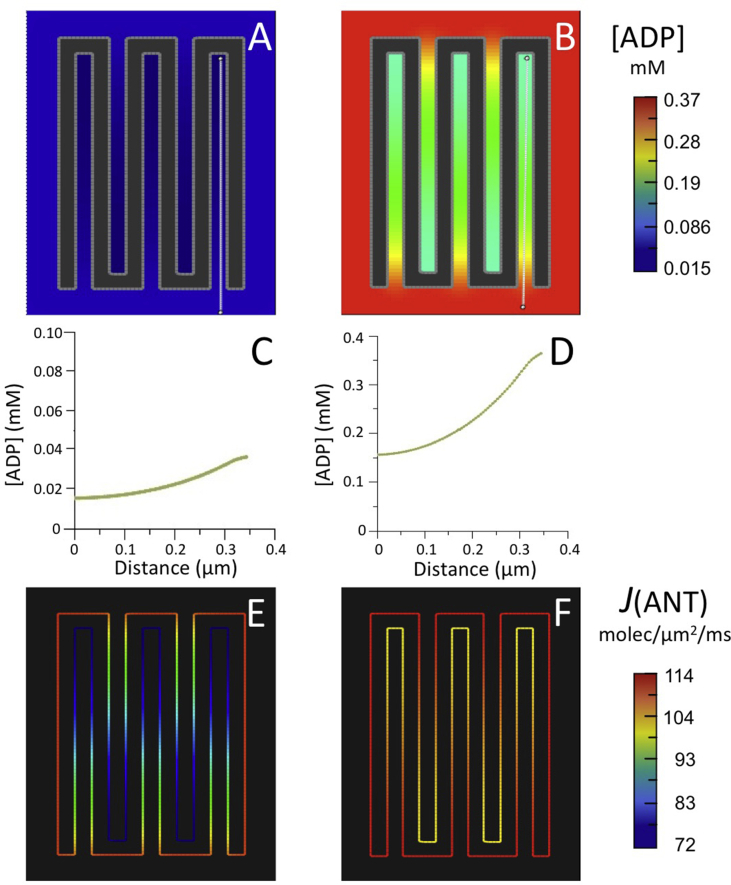
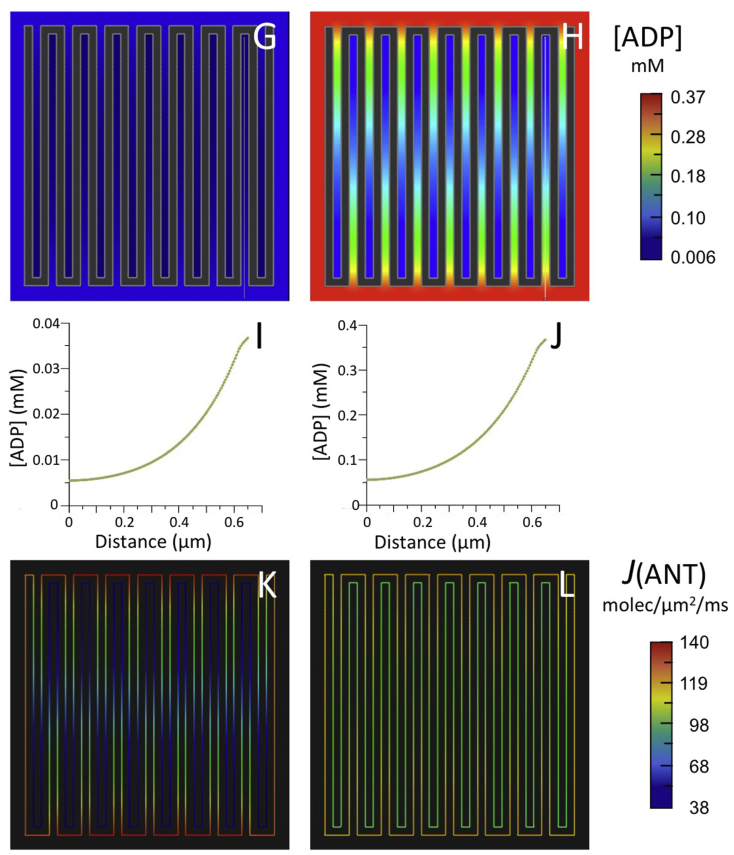
Fig. 4.
Results of computer simulations using the reduced BioModel for mitochondrial energy metabolism (Fig. 2) with (A–F) the 5-crista 2D spatial model of Fig. 3 and (G–L) a larger 15-crista 2D model, as described in text.
(A,B; G,H) Maps of steady state ADP concentration, [ADP], outside matrix for initial [ADP] of 0.037 mM (A,G) and 0.370 mM (B,H).
(C,D; I,J) Plots of [ADP] along the white lines (from distal to proximal relative to the crista junctions) in the maps of (A,B; G,H), respectively.
(E,F; K,L) Maps of ANT flux, J(ANT), along the inner membrane corresponding to the conditions of (A,B; G,H), respectively.