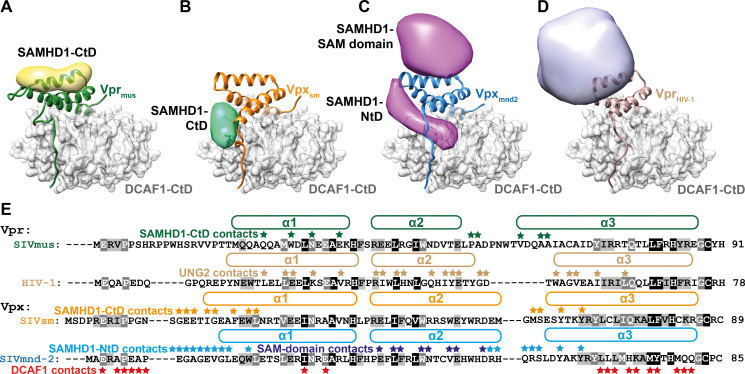
Fig 6. Variability of neo-substrate recognition in Vpx/Vpr proteins.
Comparison of neo-substrate recognition modes of Vprmus (A), Vpxsm (B), Vpxmnd2 (C) and VprHIV-1 (D). DCAF1-CtD is shown as grey cartoon and semi-transparent surface, Vprmus−green, Vpxsm−orange, Vpxmnd2 –blue and VprHIV-1– light brown are shown as cartoon. Models of the recruited ubiquitylation substrates are shown as strongly filtered, semi-transparent calculated electron density maps with the following colouring scheme: SAMHD1-CtD bound to Vprmus−yellow, SAMHD1-CtD (bound to Vpxsm, PDB 4cc9) [50]–mint green, SAMHD1-NtD (Vpxmnd2, PDB 5aja) [51]–magenta, UNG2 (VprHIV-1, PDB 5jk7) [54]–light violet. (E) Multiple sequence alignment of Vpr and Vpx proteins from A-D. Helices are indicated by the boxes above the amino acid sequences. Residues involved in neo-substrate recognition are indicated by asterisks above the amino acid sequences. Residues involved in DCAF1-binding in all Vpr and Vpx proteins are indicated by red asterisks below the Vprmnd-2 amino acid sequence. Residues shaded grey or black are at least 60% or 90% type-conserved in all Vpx and Vpr proteins, respectively.