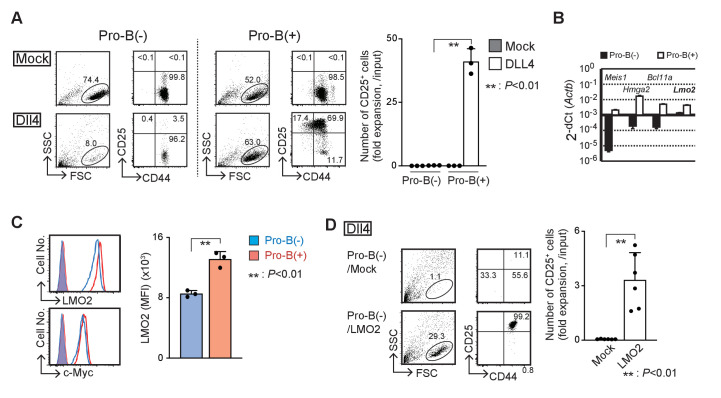
Figure 1. LMO2 is critical for the maintenance of T-cell differentiation potential in Ebf1-deficient pro-B cells.
(A) Establishment of Ebf1-deficient pro-B cell lines with or without differentiation potential to the T-cell lineage. Lineage markers (CD19, Gr1, TER119, NK1.1)-negative, c-kit-positive cells in Ebf1−/− FL were cultured on TD7 or OP9 cells, and Ebf1-deficient pro-B cell lines were established. Stably growing pro-B cells with or without T-cell potential (pro-B(+) or pro-B(−)) were cultured on OP9-Mock (Mock) or OP9-Dll4 (Dll4) cells with Flt3L, SCF, and IL7 for 6 days and analyzed for the expression of CD44 and CD25 (right panels) in the lymphoid cell gate (FSC vs. SSC, left panels) by flow cytometry. The numbers in the profiles indicate the relative percentages in each corresponding quadrant or fraction. Numbers of CD25+ cells (fold expansion/input) are shown with standard deviation (SD) (right). Statistical analysis was performed using the two-tailed Student’s t-test. **p<0.01. Data are representative of three independent experiments with similar results. (B) Reverse transcription (RT)-quantitative PCR (qPCR) detection of Meis1, Hmga2, Bcl11a, or Lmo2 transcripts in pro-B(−) (closed columns) and pro-B(+) (open columns) cells. Data represent the mean values of three independent biological replicates, and all values are normalized to the expression of Actb. Error bars indicate SD. Three independent experiments were performed, and similar results were obtained. (C) Representative intracellular staining profiles of LMO2 and c-Myc in pro-B(−) (open blue line) and pro-B(+) (open red line) cells are shown. Closed lines (orange) represent staining with control rabbit mAb of pro-B(−) and pro-B(+), which were completely merged. The average mean fluorescent intensity (MFI) of LMO2 is shown with SD (right). A two-tailed Student’s t-test was used for statistical analysis. **p<0.01. Three independent experiments were performed with similar results. (D) Introduction of Lmo2 is sufficient to maintain the T-cell differentiation potential in pro-B cells. Empty vector- or Lmo2-transduced pro-B(−) cells (pro-B(−)/Mock or pro-B(−)/LMO2) were cultured on OP9-Dll4 for 6 days and analyzed for the expression of CD44 and CD25 (right panels) in lymphoid cell gate (left panels) and rat CD2+ (lentivirus-infected) CD45+ fraction. Numbers of CD25+ cells (relative expansion/input) are shown with SD (right). **p<0.01 by two-sided Student’s t-test. Six independent experiments were performed with similar results.