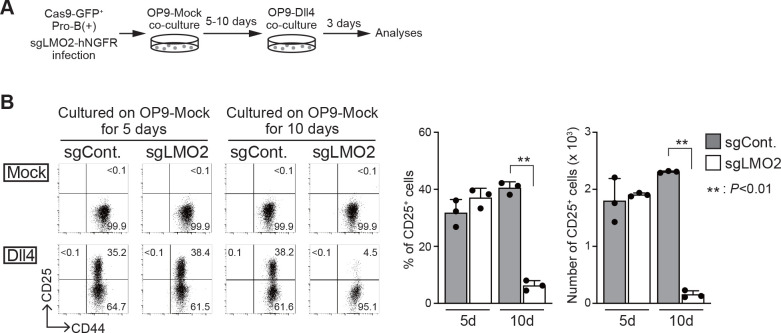
Figure 2. Loss of Lmo2 leads to the differentiation arrest in pro-B(+) cells.
(A) An experimental scheme for the deletion of Lmo2 using the CRISPR/Cas9 system in pro-B cell lines is shown. (B) Retroviral vectors encoding sgRNA against luciferase (sgCont.) or LMO2 (sgLMO2) were introduced into Cas9-expressing (GFP+) pro-B(+) cells. Five (cultured on OP9-Mock for 5 days, left panels) or 10 days (cultured on OP9-Mock for 10 days, right panels) after co-cultured on OP9-Mock cells following the infection, pro-B cells were cultured again on OP9-Mock (Mock) or OP9-Dll4 (Dll4) stromal cells for 3 days. GFP+hNGFR+ sgRNA-transduced cells were gated and analyzed for CD44 and CD25 expression (left). The percentages and numbers of CD25+ cells among GFP+hNGFR+ sgRNA-transduced cells, cultured on OP9-Dll4, are shown with SD (right). The data represent the mean values of three independent biological replicates. Each value is indicated by a closed circle. **p<0.01 by two-sided Student’s t-test.