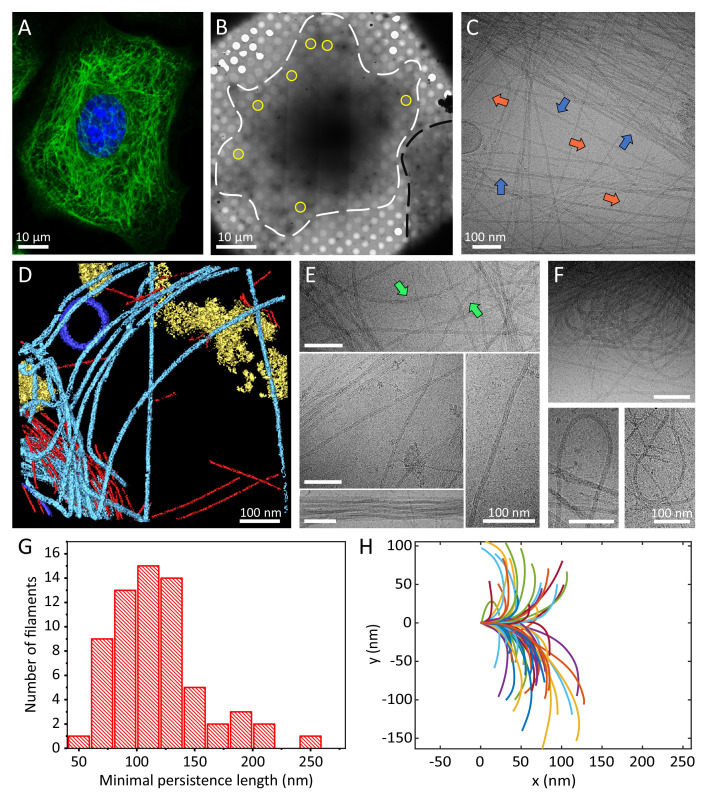
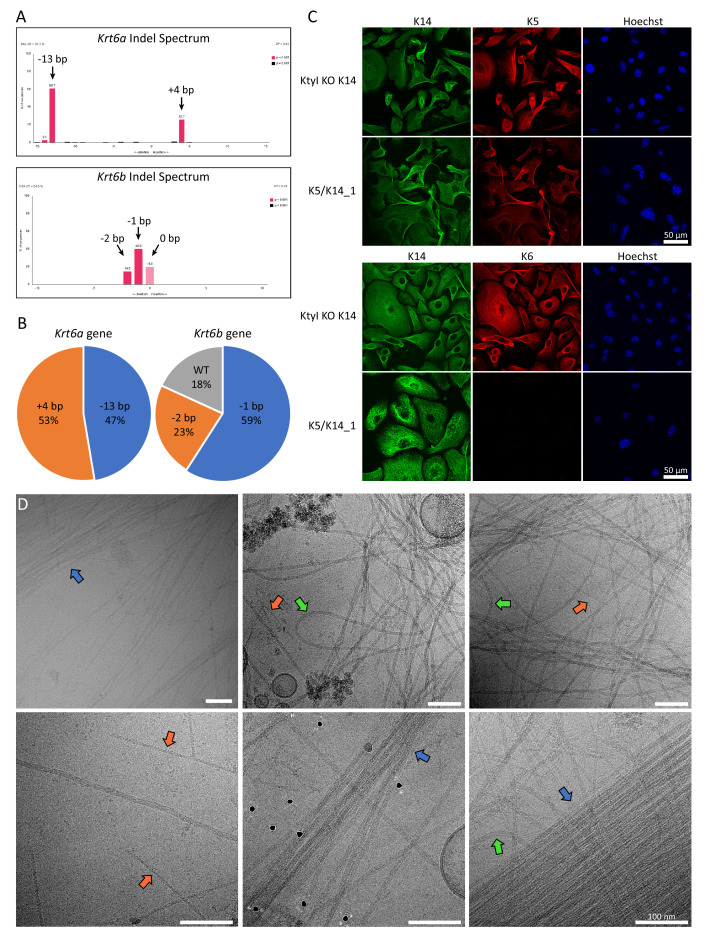
Figure 1. Cellular K5/K14 filaments as revealed by light and cryo-electron microscopy.
(A) The murine keratinocyte cell line K5/K14_1 expressing only K5 and K14 filaments forms a complex KIF meshwork, as revealed by confocal immunofluorescence. Cells were stained for K14 (green) and chromatin (blue). (B) Ghost cells were analyzed by cryo-EM and cryo-ET. Low-magnification image of a cell grown on an EM-grid and treated with detergent prior to vitrification. Cell boundaries (dashed white line) are detected as well as a neighboring cell (dashed black line). Typical regions that were analyzed by cryo-EM are marked (yellow circles). (C) A typical cryo-EM micrograph of a ghost cell imaged at a higher magnification allows the detection of keratin filaments and other cytoskeletal elements (n=1860). Keratin filaments (blue arrows) and actin filaments (orange arrows) are distinguished by their characteristic diameter. A large keratin bundle is visible in the top right corner. (D) Surface rendering view of a cryo-tomogram of a ghost cell (n=44). Keratin filaments (light blue), actin filaments (red), vesicles (dark blue), and cellular debris (yellow) were manually segmented. (E) Different organizations of keratin filaments observed in the cryo-EM micrographs (n=1860), including straight filaments (middle), curved (top, green arrows) and bundled filaments (bottom left). Scale bars: 100 nm. (F) Highly bent keratin filaments are found within cryo-EM micrographs of ghost cells. Scale bars: 100 nm. (G) Quantification of the minimal apparent persistence length measurements performed on (n=65) highly bent keratin filaments extracted from cryo-EM micrographs. (H) A plot combining 65 contours of filaments that were used for the minimal apparent persistence length measurements in (G). Individual filaments, shown in different colors, are aligned at their origins for visualization purposes.