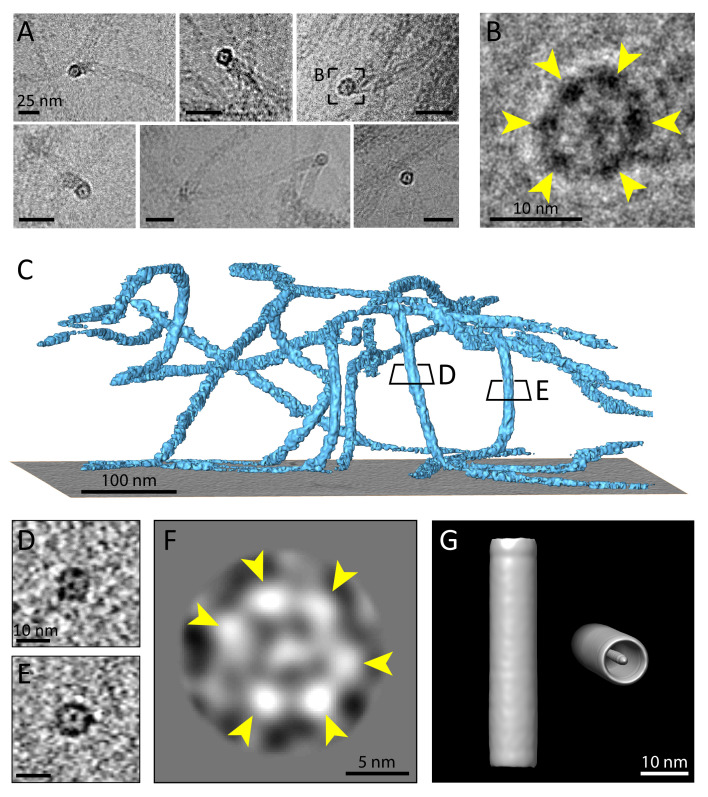
Figure 4. Multiple protofilaments and an internal electron dense core are canonical components of keratin filaments.
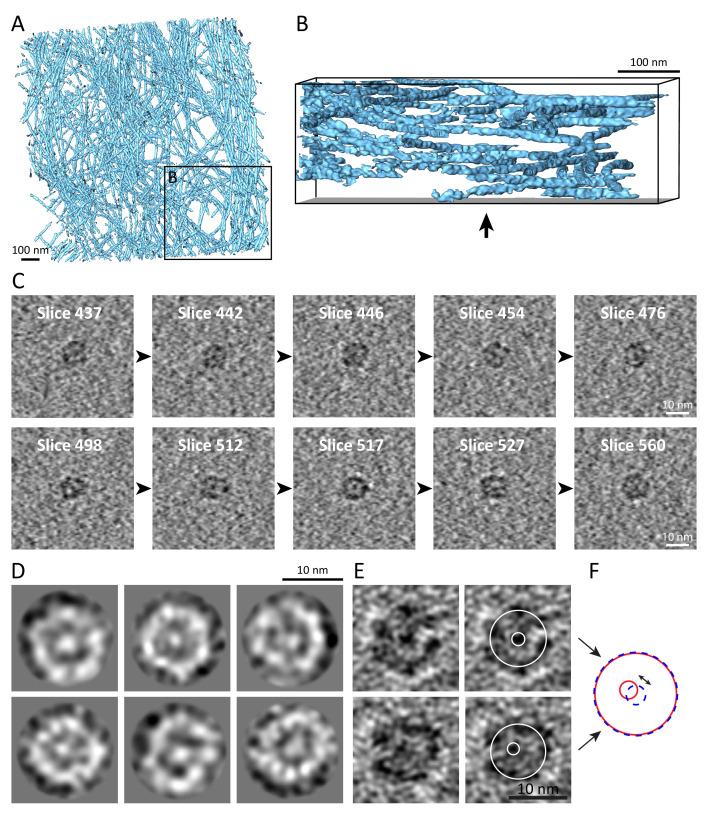
(A) Cross-section views of keratin filaments detected within the cryo-EM micrographs of ghost cells. An electron dense core is visible in the center of the keratin tube. Scale bars: 25 nm. (B) Zoomed-in view of the area boxed in (A). The cross section view reveals an internal core surrounded by six protofilaments as constituents of the tube (yellow arrowheads). (C) A surface rendered tomogram of a ghost cell was rotated in order to show the modulation of the keratin filaments within the ice layer (n=44). The three-dimensional keratin network is visualized (light blue). The level of the support is shown as a gray colored slice. Tomographic slices through vertically oriented filaments showing cross section views are indicated by boxes. (D) - (E) 7 nm thick xy-slices of the areas indicated in (C), showing KIFs as tube-like structures with a central density. Individual protofilaments can be identified. Scale bars: 10 nm. (F) A 2D class average of cross-section views extracted from individual regions of vertically oriented filaments (n=19), revealing the six individual protofilaments constituting the keratin filament tube (yellow arrowheads). (G) Low-resolution 3D model indicating the overall dimensions of a keratin filament and the presence of the central density. The structure was calculated template-free by randomizing the rotation angle of extracted 55 nm long keratin segments. Left: Side view. Right: Tilted cross section view revealing internal electron dense core.