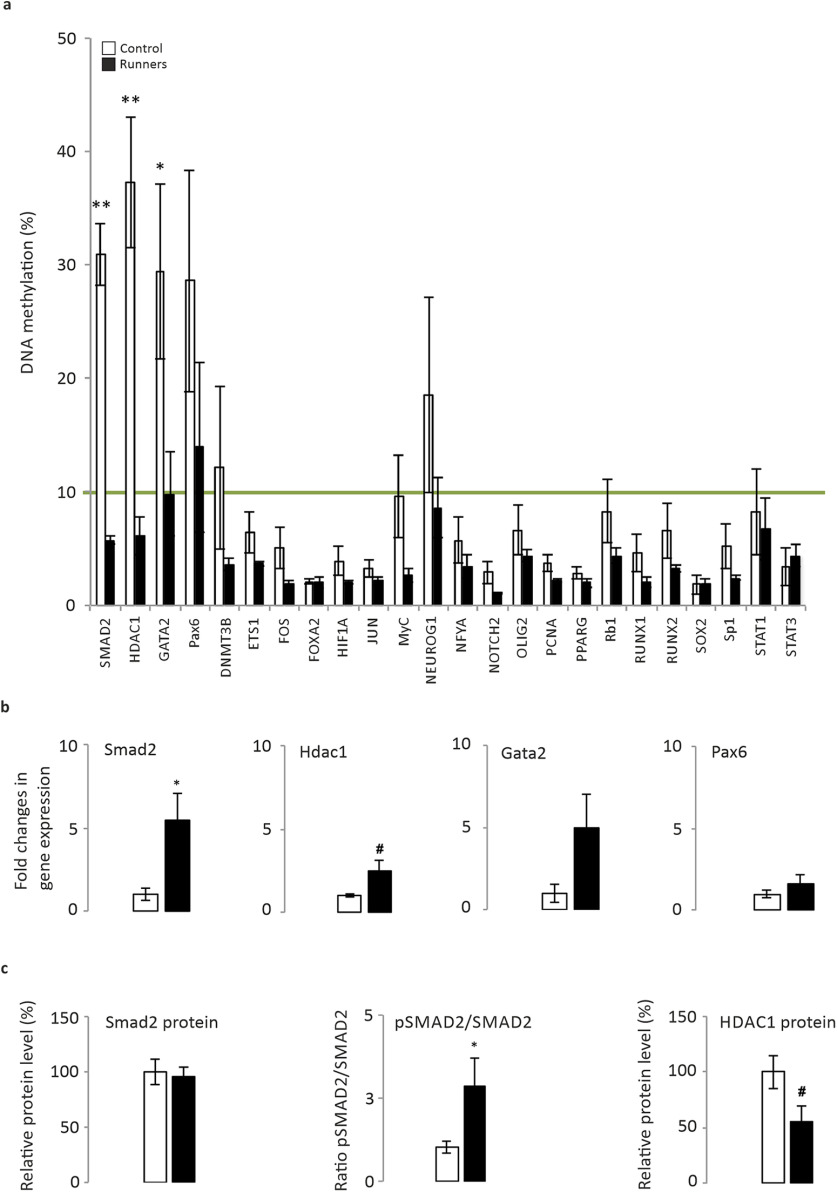
Figure 2.
Epigenetic control of gene expression in sedentary and exercising animals. a, DNA methylation array. The DNA methylation profile of 24 stem cell transcription factors was defined in the hippocampi of sedentary and exercising mice. The green line shows the threshold from which values up to 10% are considered as hypermethylation. A statistical difference in methylation was found for Smad2 (p = 0.009), Hdac1 (p = 0.009), and Gata2 (p = 0.047) genes. Values up to 10% were considered as hypermethylation. The majority of genes studied in this experiment were hypomethylated. It is worth noting that the Smad2 gene was found in a hypermethylated status in sedentary mice, while 2 weeks of physical exercise reduced the methylation level to under the threshold value (from 30.89 ± 2.74% to 5.73 ± 0.33%). Genes such as Hdac1 and Gata2 showed a methylation profile similar to those of Smad2 (Hdac1: from 37.28 ± 5.75% to 6.09 ± 1.68%; Gata2: from 29.40 ±7.67% to 9.77 ± 3.71%). b, Quantification by qPCR of mRNA expression with the most relevant changes in methylation. c, Smad2 protein quantification in Western blots, the pSmad2/total Smad2 ratio, and the HDAC1 protein. N = 5 animals/group. #0.1 > p ≥ 0.05. *p < 0.05, **p < 0.01