Abstract
Objectives
The presence of SARS-CoV-2 virus in the saliva of patients infected with COVID-19 has been confirmed by several studies. However, the use of saliva for the diagnosis of COVID-19 remains limited, because of the discrepancies in the results, which might be due to using different saliva sampling methods. The purpose of this study was to compare the consistency of SARS-CoV-2 detection using two different saliva sampling methods (oral swab and unstimulated saliva) to that of the standard nasopharyngeal swab.
Methods
Fifty-five subjects were recruited from a pool of COVID-19 inpatient at the Hospital Israelita Albert Einstein (HIAE), Brazil. Nasopharyngeal swab, oral swab, and self-collected unstimulated saliva samples were examined for SARS-CoV-2 using RT-PCR.
Results
Self-collected unstimulated saliva demonstrated 87.3% agreement in the detection of SARS-CoV-2 virus as compared with the nasopharyngeal swab, while oral swab displayed 65.9% agreement when compared to nasopharyngeal swab and 73% when compared to self-collected unstimulated saliva.
Conclusion
Unstimulated self-collected saliva samples have shown a higher agreement with the nasopharyngeal swab samples for SARS-COV-2 detection than that obtained when using oral swab samples.
Clinical relevance
This study compares the accuracy of COVID-19 test using different saliva sampling methods to that of nasopharyngeal swab. Given the need for a simple self-applied test that can be performed at home, our findings support the efficacy of self-collected unstimulated saliva samples in the diagnosis of SARS-CoV-2 infection, alleviating the demands for swab supplies, personal protective equipment, and healthcare personnel.
Keywords: Coronavirus, Coronavirus infections, SARS-CoV-2, COVID-19, Saliva, Biomarkers
Introduction
Population-level employable rapid testing is part of a strategy, where testing should be widely available, preferably outside of healthcare or hospital settings, avoiding the overload of resources and the risk of transmission to other patients and healthcare professionals [1]. To date, the gold standard for the diagnosis of SARS-CoV-2 infection is reverse transcription-polymerase chain reaction (RT-PCR) analysis of respiratory tract swabs [2]. However, oropharyngeal and nasopharyngeal collection requires trained professionals to be performed and is associated with discomfort and inconvenience to the patients, in addition to increasing risk of virus transmission, thereby halting the possibility of employing this test in a large population setting [3–5]. Therefore, it is rather imperative to design a convenient and easily accessible test to control the spread of a highly infectious disease such as COVID-19 before vaccines or anti-viral treatment become widely available.
Saliva specimen collection is one of the simplest ways to collect body fluids and represents an important source for the diagnosis and monitoring of coronavirus infection [6]. Saliva sampling has the advantage of being safe, easy to perform, and noninvasive, and may be self-collected repeatedly without discomfort to the patient. Several studies have confirmed a reliable detection of SARS-CoV-2 in the saliva of patients with COVID-19. However, the application of saliva testing for COVID-19 has been hampered due to discrepancies in the reported reliability of the test results. It is highly possible that the method of saliva sample collection could potentially be the source of the observed discrepancies in the reliability of the test results. Regarding the sampling methods, saliva samples used for COVID-19 testing have been collected in different ways, oral swab and unstimulated saliva [6]. To the best of our knowledge, there is no study that directly compared the accuracy of these two methods of sampling to that of the standard nasopharyngeal swab.
Therefore, the aim of this study was to compare the consistency of SARS-CoV-2 detection using saliva samples (oral swab and self-collected unstimulated saliva) to that of standard nasopharyngeal swab.
Methods
Ethics
The study protocol was approved by the National Ethics in Human Research Committee of Brazil (CONEP, protocol #4172–20). All patients were individually informed about the nature of the proposed study and its risks and benefits, and signed informed consent forms.
Subject population and selection criteria
COVID-19 subjects aged 18–90 years were recruited from May 2020 to June 2020 from a pool of COVID-19 inpatients at the Hospital Israelita Albert Einstein (HIAE), Brazil. Inclusion criteria were patients with positive SARS-CoV-2 determined by the nasopharyngeal/oropharyngeal swab RT-PCR method and recorded in the medical record, within 3 days of hospitalization. Excluded were pediatric patients, patients with a negative RT-PCR result for SARS-CoV-2 at the time of recruitment, and patients who had other clinical illness that in the opinion of the investigators could interfere with compliance to sample collection.
Sample collection
The collection of nasopharyngeal and saliva samples was performed within 3 days of hospitalization. Senior nurse staff took nasopharyngeal samples using a flexible, mini-tip swab, which was passed through the patient’s nostril until the posterior nasopharynx was reached, left in place for several seconds to absorb secretions then slowly removed while rotating. The swab was placed in a sterile viral transport medium (total volume 3 mL) and sealed securely. Unstimulated whole saliva was self-collected by the patients in a sterile 50-mL tube as previously described [7]. Oral swabs were taken from the back of the tongue, deep groove and floor of the mouth by a trained dentist using a flexible mini-tip swab and placed in the sterile viral transport medium (total volume 3 mL). All samples were immediately transported on ice to the laboratory at the Hospital Israelita Albert Einstein and tested within 48 h of collection [8, 9].
Detection and quantification of viral RNA
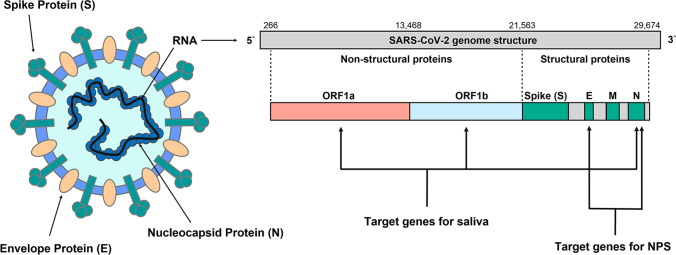
Viral RNA was extracted from 200 μL of viral transport medium from the nasopharyngeal swab or from the oral swab or from unstimulated saliva specimens using a QIAsymphony™ RNA Kit (QIAGEN, Hilden, Germany) according to the manufacturer’s instructions. Following extraction, RNA was eluted with 60 µL of elution buffer. For SARS-CoV-2 RNA detection in saliva (unstimulated saliva and oral swab), RT-qPCR amplification of the Nucleocapsid (N) and Open reading frame (ORF) 1ab genes was performed using the XGen Master COVID-19 RT-PCR Kit (Mobius Life Science Comércio de Produto para Laboratórios Ltda, Pinhais, Brazil). The sensitivity of the reaction in routine diagnosis varies according to the gene analyzed, as follows: SARS-CoV-2 gene ORF1ab gene—10 copies/reaction; SARS-CoV-2 N gene—50 copies/reaction. Saliva samples were classified as positive for SARS-CoV-2 when the cycle threshold (CT) for both N and ORF1ab genes was lower than 38. For SARS-CoV-2 RNA detection in nasopharyngeal swab specimen (NPS), RT-qPCR amplification of the Envelope (E) and N genes was performed as previously described [10]. NPS samples were classified as positive for SARS-CoV-2 when the CT for both E and N genes was lower than 38 (Fig. 1).
Fig. 1.
SARS-COV-2 diagnostic targets: The figure illustrates diagnostic targets for COVID-19 testing. Genes targets used for saliva and for nasopharyngeal swab specimen (NPS) are illustrated
Statistical analysis
The results were presented by estimated mean values, 95% confidence intervals (95% CI) and p-values. The CT measurements obtained in saliva collection were compared to the CT measurements in nasopharyngeal swab collection using models of generalized estimation equations (GEE) with gamma distribution, considering the relationship between the different collections performed in the same patient. The same calculation was performed to compare the CT measurements obtained from unstimulated saliva, oral swab, and nasopharyngeal swab collections. The sensitivity of each test (self-collected unstimulated saliva and oral swab) was determined by the proportion of positive results to that of the gold standard (nasopharyngeal swab). All analyses were performed with the SPSS program (IBM Corp, IBM SPSS Statistics for Windows, Version 24.0, Armonk, NY) considering a 5% significance level.
Results
A total of 55 patients with positive COVID-19 RT-PCR using nasopharyngeal swab were included in the study. Of the 55 patients, 48 (87.3%) were also positive with self-collected unstimulated saliva. Forty-one patients also provided oral swab samples, and of those, 27 patients (65.9%) were positive for SARS-CoV-2 RNA detection (Table 1).
Table 1.
SARS-CoV-2 virus detection in self-collected unstimulated saliva, oral swab, and nasopharyngeal swab specimens
| Specimen | Result | |
|---|---|---|
| Negative n (%) |
Positive n (%) |
|
| Unstimulated saliva (n = 55) | 7 (12.7) | 48 (87.3) |
| Oral swab (n = 41) | 14 (34.1) | 27 (65.9) |
| Nasopharyngeal swab (n = 55) | 0 (0.0) | 55 (100.0) |
Unstimulated self-collected saliva and nasopharyngeal swab specimens in the detection of SARS-CoV-2
In the RT-qPCR of saliva samples, two genes from SARS-CoV-2 were amplified, ORF1ab and N genes. Amplification of N gene was observed in all saliva samples while amplification of ORF1ab, only in 72.8%. Table 2 shows the distribution of CT by the N and ORF genes in unstimulated saliva compared to the E and N genes in nasopharyngeal swab.
Table 2.
Cycle threshold (CT) mean values and the 95% confidence interval (95% CI) comparisons for (A) N gene from self-collected unstimulated saliva and for the E and N genes from nasopharyngeal swab specimens (NPS) (n = 34) and (B) ORF gene from self-collected unstimulated saliva and for the E and N genes from NPS (n = 24)
| Unstimulated saliva | Nasopharyngeal swab | ||
|---|---|---|---|
| A | Gene N | Gene E | Gene N |
|
Mean CT 95% CI |
29.9 28.4; 31.6 |
25.8 23.5; 28.3 |
28.4 26.0; 30.9 |
| B | Gene ORF | Gene E | Gene N |
|
Mean CT 95% CI |
27.5 25.4; 29.7 |
24.1 21.5; 27.0 |
26.4 23.9; 29.1 |
Values expressed by estimated means and 95% confidence intervals (95% CI)
We observed differences between the CT for N gene in saliva samples (CT NS) and the CT for gene E (CT E) in nasopharyngeal swab specimens, where the mean CT E was − 4.1 (95%CI: − 6.8 to − 1.5; p = 0.001). However, there was no statistical difference between CT for N gene (CT N) from self-collected unstimulated saliva specimens and nasopharyngeal swab (p = 0.183). For the nasopharyngeal swab, there was no statistical difference between the CT E (p = 0.080) and CT N (p = 0.496) values (Table 3).
Table 3.
Differences between cycle thresholds obtained for the N (CT NS) and ORF (CT ORF) genes in self-collected unstimulated saliva and for the E (CT E) and N (CT NNP) genes in nasopharyngeal swab (NPS) specimens
| Sample | CT difference | Estimated mean difference (95% CI) | p-value |
|---|---|---|---|
| Saliva | CT E − CT NS | − 4.1 (− 6.8; − 1.5) | 0.001 |
| CT NNP − CT NS | − 1.6 (− 3.9; 0.7) | 0.183 | |
| NPS | CT E − CT ORF | − 3.3 (− 7.0; 0.3) | 0.080 |
| CT NNP − CT ORF | − 1.1 (− 4.1; 2.0) | 0.496 |
All values are expressed by estimated means, with 95% CIs in parentheses
Self-collected unstimulated saliva, oral swab, and nasopharyngeal swab specimens in the detection of SARS-CoV-2
Of the 55 patients, 21 had concomitantly CT values for E and N genes in nasopharyngeal swab specimens and for N gene in unstimulated saliva and oral swab specimens and 12 had concomitantly CT values for E and N genes in nasopharyngeal swab specimens and for ORF gene in unstimulated saliva and oral swab specimens. Table 4 shows the distribution of CT by the N and ORF genes in unstimulated saliva and oral swab and by the E and N (NNP) genes in nasopharyngeal swab.
Table 4.
Mean values and 95% confidence intervals (95% CI) estimated for cycle threshold (CT) measurements for (A) N gene in unstimulated saliva and oral swab and by the E and N genes in nasopharyngeal swab specimens (NPS) (n = 21) and (B) ORF gene in unstimulated saliva and oral swab and by the E and N genes in NPS (n = 12)
| Sample | ||||
|---|---|---|---|---|
| Unstimulated saliva | Oral swab | Nasopharyngeal swab | Nasopharyngeal swab | |
| A | Gene N | Gene N | Gene E | Gene N |
|
Mean CT 95% CI |
28.9 26.8; 31.1 |
33.3 31.5; 35.1 |
24.8 22.3; 27.5 |
27.7 25.0; 30.7 |
| B | Gene ORF | Gene ORF | Gene E | Gene N |
|
Mean CT 95% CI |
24.3 21.8; 27.1 |
32.7 29.4; 36.3 |
23.4 20.3; 26.8 |
25.7 22.7; 29.2 |
Values expressed by estimated means and 95% confidence intervals (95% CI)
When comparing the CT values of the N gene in the oral swab to the CT values in the nasopharyngeal specimens, we observed differences in relation to the CT E (p < 0.001) and CT NNP, where the mean CT E was − 8.5 (95% CI: − 12.2 to − 4.7 and the mean CT NNP was − 5.6 (95% CI: − 9.2 to − 1.9; p = 0.001) lower than the mean CT N in oral swab.
As for CT values for N gene, the mean CT in unstimulated saliva was − 4.4 (95% CI: − 6.3 to − 2.4; p < 0.001) lower than that in the oral swab. Comparing the CTs of the ORF gene in the oral swab to the CTs of the nasopharyngeal specimens, we observed evidence of differences in relation to the CT E and CT NNP, where the mean CT E was − 9.3 (95% CI: − 14.7 to − 3.9; p < 0.001) and the mean CT NNP was − 6.9 (95% CI: − 12.1 to − 1.8; p = 0.004) less than the mean of the CT ORF values in oral swab. Similarly, the mean CT value of the ORF gene in unstimulated saliva was − 8.3 (95% CI: − 11.5 to − 5.2; p < 0.001) less than the average of the value in the oral swab (Table 5).
Table 5.
Differences between cycle threshold (CT) measurements obtained for the N and ORF genes in self-collected unstimulated saliva and oral swab and for the E and N genes in nasopharyngeal swab specimens (n = 21)
| CT gene differences (sample source) | Estimated mean difference (95% CI) | p-value |
|---|---|---|
| CT E (NPS) − CT N (S) | − 4.1 (− 7.0; − 1.2) | 0.003 |
| CT N (NPS) − CT N (S) | − 1.2 (− 3.8; 1.5) | 0.384 |
| CT E (NPS) − CT N (OS) | − 8.5 (− 12.2; − 4.7) | < 0.001 |
| CT N (NPS) − CT N (OS) | − 5.6 (− 9.2; − 1.9) | 0.001 |
| CT N (S) − CT N (OS) | − 4.4 (− 6.3; − 2.4) | < 0.001 |
| CT E (NPS) − CT ORF (S) | − 0.9 (− 3.7; 1.8) | 0.594 |
| CT N (NPS) − CT ORF (S) | 1.4 (− 1.6; 4.4) | 0.594 |
| CT E (NPS) − CT ORF (OS) | − 9.3 (− 14.7; − 3.9) | < 0.001 |
| CT N2 (NPS) − CT ORF (OS) | − 6.9 (− 12.1; − 1.8) | 0.004 |
| CT ORF (S) − CT ORF (OS) | − 8.3 (− 11.5; − 5.2) | < 0.001 |
All values are expressed by estimated means, with 95% CIs in parentheses. CT E, CT value for E gene; CT N, CT value for N gene; CT ORF, CT value for ORF gene. Sample sources are indicated in parentheses as follows: (NPS), nasopharyngeal swab specimen; (S), saliva; (OS), oral swab
Types of saliva sampling for SARS-CoV-2 detection
When comparing unstimulated self-collected saliva and oral swab, of the 37 patients with positive results for gene N in self-collected unstimulated saliva, 26 (70.3%) were also positive with the oral swab. Considering the positive results for the ORF gene in unstimulated saliva in 34 patients, 16 (47.1%) were also positive with the oral swab. Regardless of the gene, unstimulated saliva was positive in 37 patients, whereas the oral swab was positive only in 27 of the 37 (73.0%) samples.
Discussion
Saliva samples can be collected by a variety of methods, such as posterior oropharyngeal saliva, oral swab, and unstimulated saliva. However, the majority of studies used the term saliva failing to describe exactly which sample collection technique was used [6]. In the present study, we directly compared two saliva sampling techniques for SARS-CoV-2 detection. While self-collected unstimulated saliva showed a sensitivity of 87.3% when compared to nasopharyngeal swab, this result was decreased to 65.9% when the saliva samples were collected using oral swab. This finding is relevant in the current pandemic, as the use of a swab for saliva collection does not seem optimal. Using self-collected unstimulated saliva as the sampling method alleviates the need for swabs and healthcare workers. Thus, self-collection of saliva in simple, sterile, nuclease-free tubes, negating the high costs associated with specialized collection devices, could be one of the key obstacles to meet the mass testing demands [11].
Here, we have demonstrated that nasopharyngeal swab exhibits higher sensitivity for COVID-19 testing than unstimulated self-collected saliva. However, unstimulated self-collected saliva showed higher sensitivity than oral swab. While nasopharyngeal swab samples appear to be more suitable for COVID-19 testing, it is important to recognize that saliva-based COVID-19 testing is an important option for frequent/repeated testing [12], as being scalable, and provides a useful sustainable strategy that easily allows periodic testing over time [13].
The sensitivity of self-collected unstimulated saliva was reduced to 78.2% when using only results from ORF gene. Although RT-PCR has been used as the gold standard for diagnosing COVID-19, there are some difficulties involving its application [14]. The first RT-PCR kit made available by China’s CDC has as targets the N and ORF1ab genes of SARS-CoV-2 genome, and the infection is confirmed when both genes are amplified. However, there are reports of the amplification of only one of these gene targets [15].
Previous studies have reported a tendency for a higher viral load (or lower CTs) in nasopharyngeal swabs in comparison to saliva samples [16, 17]. However, in the present study, we found no significant differences between CT N in nasopharyngeal swab and in unstimulated saliva specimens (p = 0.183). This was not true when comparing CT N of nasopharyngeal swab to oral swab, where the latter presented a higher CT. The sensitivity of different diagnostic methods may vary according to variables such as gene target, sample processing and clinical signs and symptoms at the time of sample collection, as observed in the present study.
Recent studies reported that SARS-CoV-2 could be detected in the saliva of asymptomatic individuals and outpatients [17–20], showing a higher sensitivity than nasopharyngeal swabs for diagnosis of asymptomatic and mild COVID-19 infection [21–23]. In the present study, we have only included inpatients, although we see great potential with saliva’s utility outside of the hospital setting.
There are, of course, limitations in the present study. Primarily, only symptomatic inpatients testing positive for COVID-19 [24] were included in the current investigation. Furthermore, as molecular biology supplies were limited, two different COVID-19 RT-PCR kits were used. We also recognize the limited number of study participants who were able to provide all three different samples. Outside of a pandemic setting, where there are no limitations of resources, our future studies will include a larger group of patients, record the operational characteristics (sensitivity, specificity, precision, and so on) of COVID-19 diagnostic test, and comply with Standards for Reporting of Diagnostic Accuracy Studies (STARD) guidelines [25]. Finally, no biological tests have 100% sensitivity and 100% specificity, a fact which needs to be considered when diagnostic results are translated into clinical practice [26].
Future investigations should include a larger sample size and focus on the diagnostic capability of human saliva for identifying COVID-19 with new cost-effective point-of-care technologies, making diagnosis accessible for anyone and anywhere [27]. Self-collected saliva is quite comfortable for patients as well as being easy, low cost [28], and non-invasive with minimal equipment, and does not require a healthcare facility or professionals. It also minimizes the nosocomial transmission of SARS-CoV-2 to healthcare workers. In the current pandemic situation, all research centers, health agencies, and healthcare providers should explore this diagnostic opportunity and rapidly develop automated molecular point-of-care assays.
Conclusions
Unstimulated self-collected saliva samples have shown a higher agreement with the nasopharyngeal swab samples for SARS-COV-2 detection than that obtain with oral swab samples. Self-collection of unstimulated saliva samples is safe and alleviates demands for supplies of swabs, personal protective equipment, and healthcare personnel. Given the growing need for testing worldwide, our findings support the efficacy of saliva samples in the diagnosis of SARS-CoV-2 infection.
Acknowledgements
We are grateful for the study participants for their time and commitment to the study. We would like to thank all members of the clinical diagnosis laboratory at the Hospital Israelita Albert Einstein for their dedication and work, which made this study possible. The authors thank Dr. Fernanda Mansur for editing the manuscript and her help with graphic design. Additional support in the statistical analysis was provided by Sandra Regina Malagutti of the Hospital Israelita Albert Einstein.
Funding
The work was supported by Colgate-Palmolive Company, USA, and the Latin American Oral Health Association.
Declarations
Ethics approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all individual participants included in the study.
All authors read and agreed on the final version of the manuscript.
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Fernanda de Paula Eduardo, Leticia Bezinelli and Debora Heller contributed equally.
References
- 1.Salathé M, Althaus CL, Neher R, et al. COVID-19 epidemic in Switzerland: on the importance of testing, contact tracing and isolation. Swiss Med Wkly. 2020;150:w20225. doi: 10.4414/smw.2020.20225. [DOI] [PubMed] [Google Scholar]
- 2.Jin Y, Wang M, Zuo Z, Fan C, Ye F, Cai Z, Wang Y, Cui H, Pan K, Xu A. Diagnostic value and dynamic variance of serum antibody in coronavirus disease. Int J Infect Dis. 2020;94:49–52. doi: 10.1016/j.ijid.2020.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.To KK-W, Tsang OT-Y, Yip CC-Y, et al. Consistent detection of 2019 novel coronavirus in saliva. Clin Infect Dis. 2020;71(15):841–843. doi: 10.1093/cid/ciaa149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Azzi L, Carcano G, Gianfagna F, et al. Saliva is a reliable tool to detect SARS-CoV-2. J Infect. 2020;81(1):e45–e50. doi: 10.1016/j.jinf.2020.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.To KK, Tsang OT, Leung WS, et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. 2020;20(5):565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fernandes LL, Pacheco VB, Borges L, et al. Saliva in the diagnosis of COVID-19: a review and new research directions. J Dent Res. 2020;99(13):1435–1443. doi: 10.1177/0022034520960070. [DOI] [PubMed] [Google Scholar]
- 7.Thomadaki K, Bosch J, Oppenheim F, Helmerhorst E. The diagnostic potential of salivary protease activities in periodontal health and disease. Oral Dis. 2013;19(8):781–788. doi: 10.1111/odi.12069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Griesemer SB, Van Slyke G, Ehrbar D, Strle K, Yildirim T, Centurioni DA, Walsh AC, Chang AK, Waxman MJ, George KS. Evaluation of specimen types and saliva stabilization solutions for SARS-CoV-2 testing. Infectious Diseases (except HIV/AIDS) medRxiv [Preprint] 2020 doi: 10.1101/2020.06.16.20133041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ott I, Vogels C, Grubaugh N, Wyllie A (2020) Saliva collection and RNA extraction for SARS-CoV-2 detection. Available from: 10.17504/protocols.io.bh6mj9c6
- 10.Corman VM, Landt O, Kaiser M et al (2020) Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill 25(3). 10.2807/1560-7917.ES.2020.25.3.2000045 [DOI] [PMC free article] [PubMed]
- 11.Ott IM, Strine MS, Watkins AE, et al. Simply saliva: stability of SARS-CoV-2 detection negates the need for expensive collection devices. medRxiv [Preprint] 2020 doi: 10.1101/2020.08.03.20165233. [DOI] [Google Scholar]
- 12.Savela ES, Winnett A, Romano AE et al (2021) SARS-CoV-2 is detectable using sensitive RNA saliva testing days before viral load reaches detection range of low-sensitivity nasal swab tests. medRxiv [Preprint]. 2021 Apr 7:2021.04.02.21254771. 10.1101/2021.04.02.21254771
- 13.Tan SH, Allicock O, Armstrong-Hough M, Wyllie AL. Saliva as a gold-standard sample for SARS-CoV-2 detection. Lancet Respir Med. 2021;9(6):562–564. doi: 10.1016/S2213-2600(21)00178-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Y, Wang Y, Chen Y, Qin Q. Unique epidemiological and clinical features of the emerging 2019 novel coronavirus pneumonia (COVID-19) implicate special control measures. J Med Virol. 2020;92(6):568–576. doi: 10.1002/jmv.25748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li X, Geng M, Peng Y, Meng L, Lu S. Molecular immune pathogenesis and diagnosis of COVID-19. J Pharm Anal. 2020;10(2):102–108. doi: 10.1016/j.jpha.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sapkota D, Soland TM, Galtung HK, Sand LP, Giannecchini S, To KKW, Mendes-Correa MC, Giglio D, Hasseus B, Braz-Silva PH. COVID-19 salivary signature: diagnostic and research opportunities. J Clin Pathol. 2020;74:344–349. doi: 10.1136/jclinpath-2020-206834. [DOI] [PubMed] [Google Scholar]
- 17.Williams E, Bond K, Zhang B, Putland M, Williamson DA. Saliva as a noninvasive specimen for detection of SARS-CoV-2. J Clin Microbiol. 2020;58(8):e00776-20. doi: 10.1128/JCM.00776-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kojima N, Turner F, Slepnev V, Bacelar A, Deming L, Kodeboyina S, Klausner JD (2020) Self-collected oral fluid and nasal swab specimens demonstrate comparable sensitivity to clinician-collected nasopharyngeal swab specimens for the detection of SARS-CoV-2. Clin Infect Dis ciaa1589. 10.1093/cid/ciaa1589 [DOI] [PMC free article] [PubMed]
- 19.Pasomsub E, Watcharananan SP, Boonyawat K, Janchompoo P, Wongtabtim G, Suksuwan W, Sungkanuparph S, Phuphuakrat A. Saliva sample as a non-invasive specimen for the diagnosis of coronavirus disease 2019: a cross-sectional study. Clin Microbiol Infect. 2020;27(2):285.e1–285.e4. doi: 10.1016/j.cmi.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wyllie AL, Fournier J, Casanovas-Massana A, et al. Saliva or nasopharyngeal swab specimens for detection of SARS-CoV-2. N Engl J Med. 2020;383(13):1283–1286. doi: 10.1056/NEJMc2016359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Teo AKJ, Choudhury Y, Tan IB, et al. Saliva is more sensitive than nasopharyngeal or nasal swabs for diagnosis of asymptomatic and mild COVID-19 infection. Sci Rep. 2021;11(1):3134. doi: 10.1038/s41598-021-82787-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Savela ES, Winnett A, Romano AE et al (2021) SARS-CoV-2 is detectable using sensitive RNA saliva testing days before viral load reaches detection range of low-sensitivity nasal swab tests. Preprint. medRxiv. 2021;2021.04.02.21254771. 10.1101/2021.04.02.21254771
- 23.Tan SH, Allicock O, Armstrong-Hough M, Wyllie AL. Saliva as a gold-standard sample for SARS-CoV-2 detection. Lancet Respir Med. 2021;9(6):562–564. doi: 10.1016/S2213-2600(21)00178-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.ANVISA (2021) Nota Técnica COVID-19 Revisão 20. https://www.saude.ms.gov.br/wp-content/uploads/2021/03/Nota-Tecnica-COVID-19-Revisao-20-1.pdf. Accessed 25 May 2021
- 25.Bossuyt PM, Reitsma JB, Bruns DE, et al. STARD 2015: an updated list of essential items for reporting diagnostic accuracy studies. Radiology. 2015;277(3):826–832. doi: 10.1148/radiol.2015151516. [DOI] [PubMed] [Google Scholar]
- 26.Vandenberg O, Martiny D, Rochas O, et al. Considerations for diagnostic COVID-19 tests. Nat Rev Microbiol. 2021;19:171–183. doi: 10.1038/s41579-020-00461-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khurshid Z, Asiri FYI, Wadaani HA. Human saliva: non-invasive fluid for detecting novel coronavirus (2019-nCoV) Int J Environ Res Public Health. 2020;17(7):2225. doi: 10.3390/ijerph17072225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bastos ML, Sara Perlman-Arrow S, Menzies D, Campbell JR. The sensitivity and costs of testing for SARS-CoV-2 infection with saliva versus nasopharyngeal swabs a systematic review and meta-analysis. Ann Intern Med. 2021;174(4):501–510. doi: 10.7326/M20-6569. [DOI] [PMC free article] [PubMed] [Google Scholar]