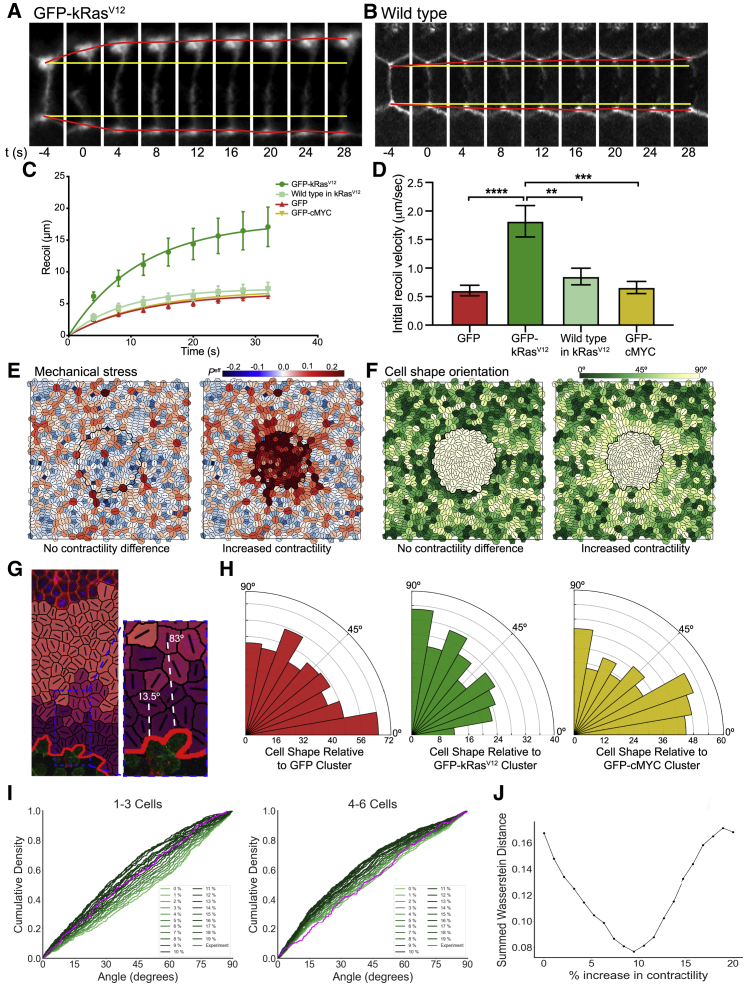
Figure 2.
kRasV12 cell cluster imposes a mechanical strain on the wild-type epithelium
(A and B) Cropped regions of confocal time-lapse stills showing laser ablation at a cell edge (highlighted by cherry-UtrCH: F-actin) in a GFP-kRasV12 cluster (A) and a surrounding wild-type cell (B). Ablation occurs at t = 0, yellow lines show the original positions of cell vertices before laser ablation, and red lines show the real-time positions of cell vertices.
(C) Recoil measurements for cells in GFP-control (red), GFP-kRasV12 (green), and GFP-cMYC (yellow) clusters and areas of wild-type tissue around GFP-kRasV12 clusters (wild type; light green); n = 10 cells for each sample; error bars are SEM.
(D) Initial recoil velocity calculated from recoil measurements in (C); one-way ANOVA: ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001; n = 10 cells for each sample; error bars are SEM.
(E) Simulated tissue, randomly generated, starting under conditions of zero net tissue stress. Heatmap indicates magnitude of cell-level isotropic stress, , with cells being under net tension (red) or compression (blue). A simulated Ras cluster was initialized in the center of the tissue (enclosed within black ring). Left: no additional contractility in cluster is shown. Right: 30% increase in cortical contractility, , in cluster is shown.
(F) Simulated tissues from (E), with heatmap showing the orientation of the principal axis of cell shape relative to the cluster (as shown in G).
(G) From confocal images, the shapes of host cells neighboring the clusters (dark purple: 1–3 cells from cluster; light purple: 4–6 cells; pink: 7+ cells) were traced and cell shape orientation (long-axis) relative to the cluster was measured (two examples in white are shown).
(H) Rose histograms showing the orientation of wild-type cells’ long axes 1–3 cells from GFP-control (red), GFP-kRasV12 (green), and GFP-cMYC (yellow) clusters, relative to the cluster, with the total number of cells analyzed across all embryos in each data group in 10° bins. Kruskal-Wallis test: GFP versus GFP-kRasV12 p < 0.01 and GFP versus GFP-cMYC p > 0.9999; n = 431 cells from 7 GFP embryos, 224 cells from 5 GFP-kRasV12 embryos, and 348 cells from 7 GFP-cMYC embryos.
(I) Cumulative distributions of cell shape orientation relative to cluster (as shown in G), from experiments (magenta) and simulations (green). Ras clusters were simulated with varying degrees of increased cortical contractility, .
(J) Wasserstein distance between experiments and simulations for cumulative distributions in (I) and Figure S2E. Discrete intervals on the x axis relate to shades of green in (I). For every contractility interval, the y axis shows the sum of the Wasserstein distances over the three distance categories (1–3, 4–6, and 7+ cells). The best fit is found at a 9% increase in contractility, where summed Wasserstein distance is minimized.
See also Figure S2.