Figure 3.
The wild-type epithelium responds to oncogene-expressing clusters with altered cell division
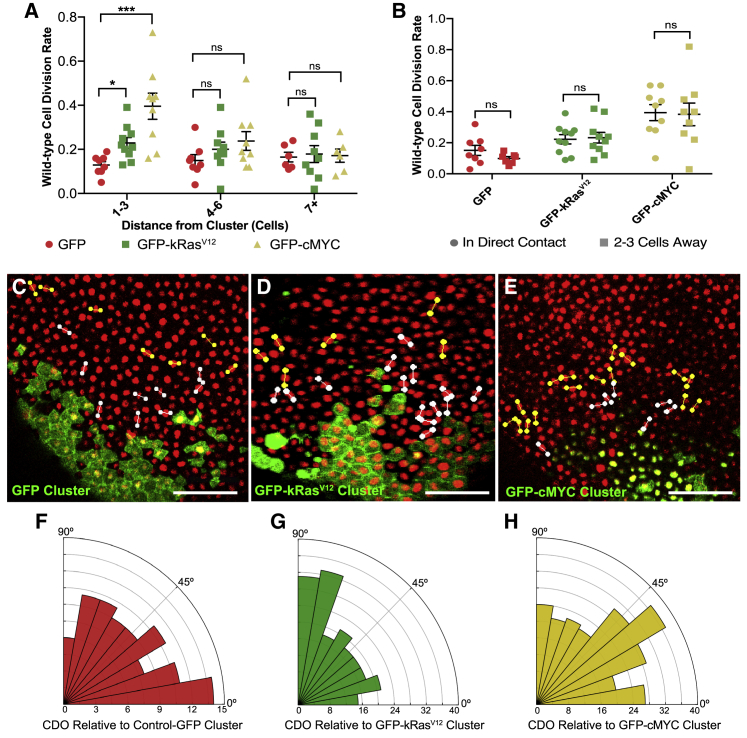
(A and B) Dot plots showing the percentage of wild-type cells that divided per minute of time lapse at different distances from GFP, GFP-kRasV12, or GFP-cMYC clusters. (A) Kruskal-Wallis test: ∗p < 0.05 and ∗∗∗p < 0.001; n = 8 GFP-control, 10 GFP-kRasV12, and 9 GFP-cMYC embryos. Error bars are SEM. (B) Paired t tests were performed; n = 8 GFP, 10 GFP-kRasV12, and 9 GFP-cMYC embryos.
(C–E) Snapshots from confocal microscopy time lapses of representative embryos showing the orientation of cell divisions that occurred in wild-type cells: colored lines were drawn, connecting the dividing anaphase nuclei, and are shown cumulatively for the entire time lapse in one snapshot. White lines label divisions 1–3 cells from the cluster, and yellow lines mark divisions 4–6 cells away. Scale bars represent 100 μm.
(F–H) Rose histograms showing cell division orientation up to 6 cells away from (F) GFP control, (G) GFP-kRasV12, and (H) GFP-cMYC clusters, with the total number of cell divisions analyzed across all embryos in each data group in 10° bins. Kruskal-Wallis test: GFP versus GFP-kRasV12 p < 0.05 and GFP versus GFP-cMYC p > 0.9999; n = 88 divisions from 8 GFP embryos, 193 divisions from 11 GFP-kRasV12 embryos, and 231 divisions from 9 GFP-cMYC embryos.
See also Figure S3.