Figure 4.
Actomyosin contraction in cluster is required to generate strain and alter cell division in wild-type tissue
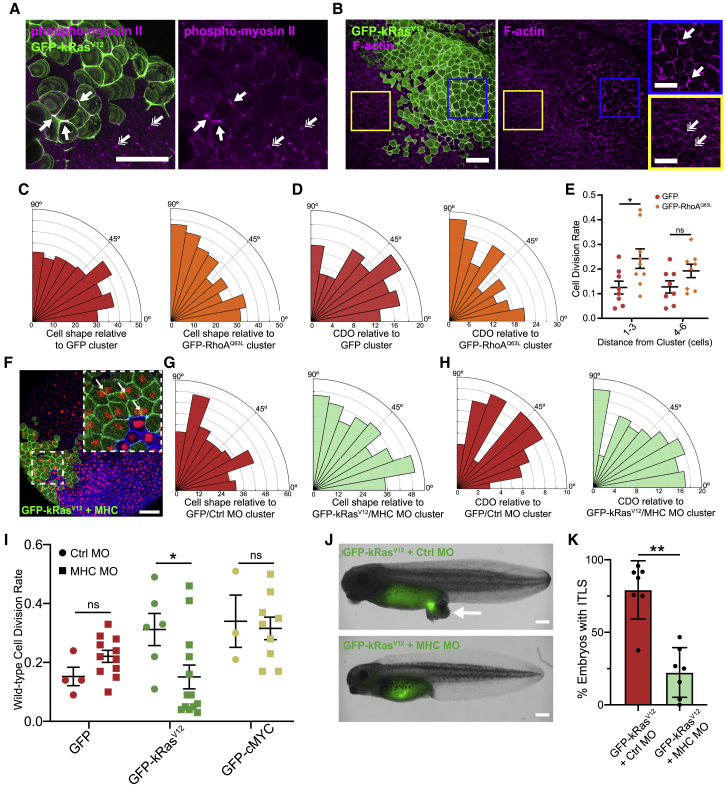
(A and B) Confocal images of fixed, stage 10 embryos with a GFP-kRasV12 cluster, stained for (A) phosphorylated myosin II (magenta), single-headed arrows highlight tricellular junctions with increased phospho-myosin II in GFP-kRasV12 cells compared to wild-type tissue (double-headed arrows), and (B) F-actin (phalloidin; magenta), single-headed arrows highlight increased F-actin at the cell cortex in the GFP-kRasV12 cluster compared to wild-type tissue (double-headed arrows).
(C) Rose histograms showing the orientation of wild-type cells’ long axes up to 6 cells from GFP-control (red) or GFP-RhoAQ63L (orange) cell clusters, relative to the cluster, in 10° bins. Kolmogorov-Smirnov test: p < 0.05; n = 298 cells from 6 GFP-control embryos and 299 cells from 6 GFP-RhoAQ63L embryos.
(D) Rose histograms showing cell division orientation relative to GFP-control (red) or GFP-RhoAQ63L (orange) clusters, with the total number of cells in 10° bins. Kolmogorov-Smirnov test: p < 0.05; n = 98 divisions from 10 GFP-control embryos and 174 divisions from 9 GFP-RhoAQ63L embryos.
(E) Dot plot showing percentage of wild-type cells that divided per minute of time lapse at different distances from GFP-control or GFP-RhoAQ63L clusters. One-way ANOVA: ∗p < 0.05; n = 7 GFP-control and 9 GFP-RhoAQ63L embryos. Error bars are SEM.
(F) Confocal microscopy image shows a myosin-II-deficient GFP-kRasV12 cell cluster. Arrows highlight “butterfly nuclei.”
(G) Rose histograms showing the orientation of wild-type cell long axes up to 3 cells from GFP/Ctrl MO (red) or myosin-II-deficient (MHC MO) GFP-kRasV12 (light green) cell clusters, in 10° bins. Kruskal-Wallis test: p > 0.9999; n = 325 cells from 6 GFP/Ctrl MO embryos and 368 cells from 7 GFP-kRasV12/MHC MO embryos.
(H) Rose histograms show cell division orientation up to 6 cells from (D) GFP/Ctrl MO (red) or GFP-kRasV12/MHC MO (light green) cell clusters, in 10° bins. Kruskal-Wallis Test: p = 0.9327; n = 58 divisions from 6 GFP/Ctrl MO embryos and 132 divisions from 9 GFP-kRasV12/MHC MO embryos.
(I) Dot plot shows percentage of wild-type cells that divided per minute of time lapse, up to 3 cells from GFP, GFP-kRasV12, or GFP-cMYC control morpholino clusters or myosin-II-deficient GFP, GFP-kRasV12, or GFP-cMYC clusters. Kruskal-Wallis test: ∗p < 0.05; n = 5 GFP/Ctrl MO embryos, 11 GFP/MHC MO, 6 GFP-kRasV12/Ctrl MO, 13 GFP-kRasV12/MHC MO, 3 GFP-cMYC/Ctrl MO, and 9 GFP-cMYC/MHC MO embryos.
(J) Images of representative embryos at stage 38 selected for presence of GFP-kRasV12 clusters at stage 10 and co-injected at 32-cell stage with Ctrl MO or MHC MO. Arrow indicates formation of ITLS in GFP-kRasV12/Ctrl MO embryo, but not GFP-kRasV12/MHC MO.
(K) Quantification of ITLS formation at stage 38 in kRasV12/Ctrl MO and GFP-kRasV12/MHC MO embryos (p < 0.01; Mann Whitney test; n = 7 independent experiments; a total of 163 GFP-kRasV12/Ctrl MO and 124 GFP-kRasV12/MHC MO embryos were assessed). Error bars are SEM.
Scale bars represent 100 μm in (A), (B) (main image), and (F); 50 μm in (B) (zoom-ins); and 500 μm in (J). See also Figure S4.