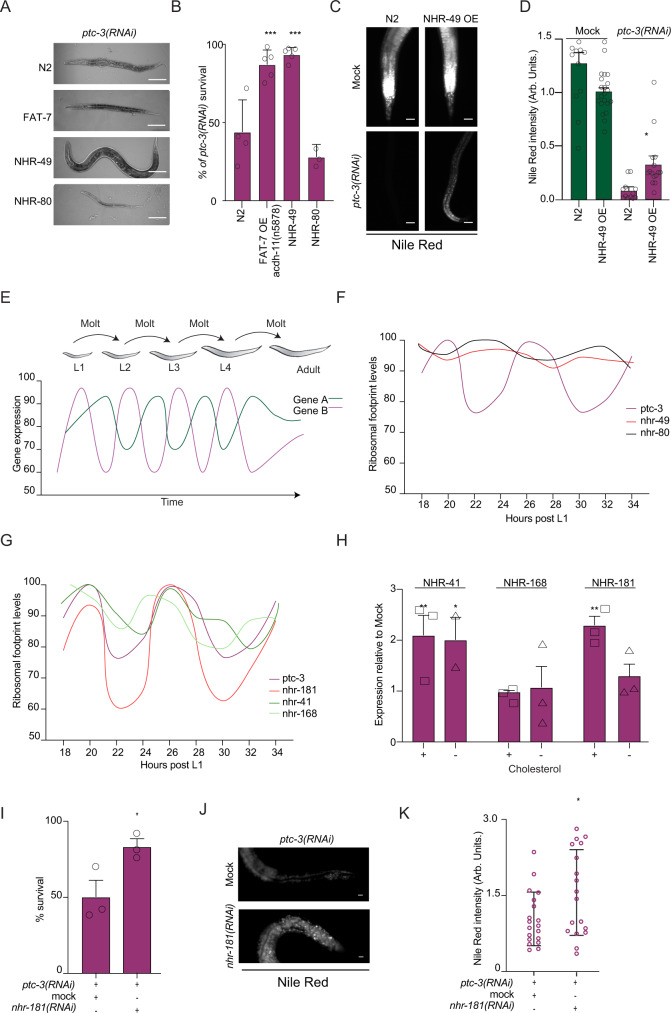
Fig. 6. PTC-3 influences NHR function in a cholesterol-dependent manner.
A Overexpression of NHR-49 or FAT-7 partially rescues ptc-3(RNAi) defects. Representative DIC images of worms. Scale bars 100 µm. B Quantification of survival rate upon overexpression of FAT-7, NHR-49 or NHR-80 in ptc-3(RNAi) animals. Mean and SD error bars. FAT-7 ***p = 0.0004, NHR-49 ***p = 0.0001. (C) Over expression of NHR-49 partially restores fat accumulation in ptc-3(RNAi) animals. Nile Red staining. Scale bars 20 µm. (D) Quantification of data shown in (C) *p = 0.0496 n = 11 (Mock N2), 21 (Mock; NHR-49 OE), 12 (ptc-3(RNAi) N2) and 15 (ptc-3(RNAi) NHR-49 OE) animals from 3 independent experiments. Mean and SEM error bars. E Schematic representation of ribosomal footprints of mRNA during C. elegans larval development. Oscillatory changes in mRNA levels during developmental time. The timing, amplitude, and whether a gene is oscillating is gene specific. F PTC-3, but not NHR-49 or NHR-80, expression oscillates during development. Data plotted from46. G Ribosomal footprint oscillations of NHR-181, NHR-168, and NHR-41 are similar to PTC-3 (data from ref. 46). (H) NHR-181 expression is modulated dependent on cholesterol levels. qRT-PCR analysis of NHR-41, NHR-168, and NHR-181 in the presence or absence of cholesterol in the growth medium. Mean and SEM error bars. NHR-41 **p = 0.004 *p = 0.0144, NHR-181 **p = 0.0011. I Genetic interaction between PTC-3 and NHR-181. Knockdown of NHR-181 rescues ptc-3(RNAi) lethality. Error bars are SEM *p = 0.0496 n = 244 (Mock; ptc-3(RNAi)) and 164 (nhr-181(RNAi); ptc-3(RNAi)) animals from three independent experiments. J nhr-181(RNAi) partially restores fat accumulation in ptc-3(RNAi) animals. Nile Red staining. Scale bars 20 µm. K Quantification of data shown in (J) *p = 0.0306 n = 19 (Mock; ptc-3(RNAi)) and 18 (nhr-181(RNAi);ptc-3(RNAi)) animals from three independent experiments. Mean and SD error bars. Statistical source data are available as source data.