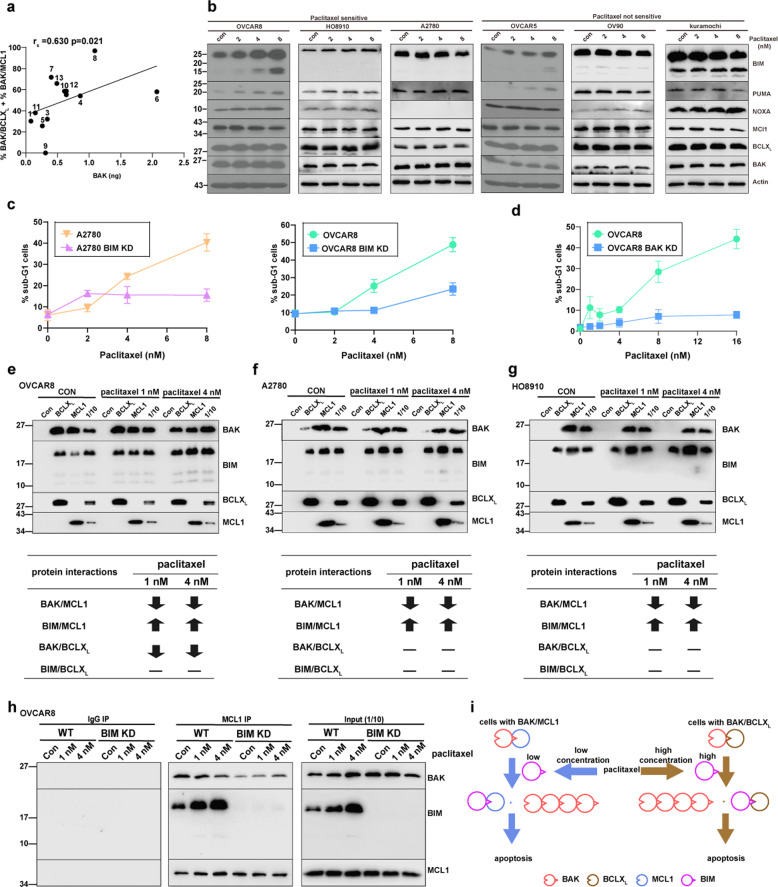
Fig. 5. BIM induced by low-dose paclitaxel binds MCL1 and disrupts the BAK/MCL1 complexes.
a Spearman correlation analysis between protein levels (ng/105 cells) of BAK and %BAK/BCLXL + %BAK/MCL1. b After paclitaxel sensitive (OVCAR8, HO8910, A2780) and paclitaxel insensitive cells (OVCAR5, OV90, Kuramochi) were treated with the indicated concentrations of paclitaxel, whole-cell lysates were blotted for indicated proteins. c, d A2780 (left panel of c), or OVCAR8 (right panel of c, and d) cells were transfected with indicated siRNA to induce protein knockdown (KD). After 24 h, cells were treated with the indicated concentrations of paclitaxel for 48 h, the percentages of sub-G1 cells were detected by flow cytometry. (n = 3 independent experiments, mean ± S.D.) e–g After OVCAR8 (e), HO8910 (f), or A2780 (g) cells were treated with the indicated concentrations of paclitaxel in the presence of Q-VD-OPh for 48 h, CHAPS lysates were immunoprecipitated with rabbit IgG (CON), or antibodies to BCLXL or MCL1. The immunoprecipitated proteins together with the indicated amount of input (1/10 or 1/4) were blotted with the indicated antibodies (upper panels) and analyzed (bottom panels). h After OVCAR8 were transfected with BIM siRNA (BIM KD) for 24 h, cells were then treated with the indicated concentrations of paclitaxel in the presence of Q-VD-OPh for 48 h. CHAPS lysates were then prepared for immunoprecipitation with rabbit IgG (CON) or antibodies to MCL1. The immunoprecipitated proteins together with indicated one-tenth of input were blotted with the indicated antibodies. i Proposed model to explain why cells with BAK/MCL1 complexes were more sensitive to paclitaxel.