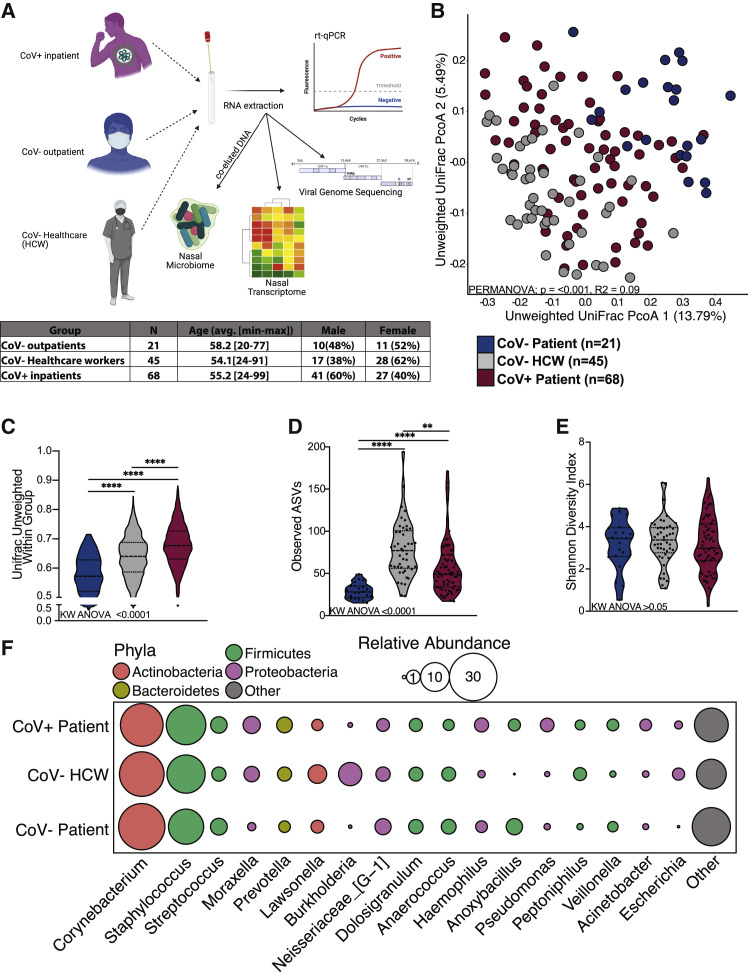
Figure 1.
The nasal microbiome of SARS-CoV-2-infected patients is distinct
(A) Study design.
(B) Principal coordinate analysis of nasal microbial communities unweighted UniFrac distance colored by host status. The contribution of host status to the total variance in the unweighted UniFrac dissimilarity was measured using permutational multivariate analysis of variance (PERMANOVA).
(C–E) Violin plot of (C) average unweighted UniFrac distances, (D) number of observed ASVs, and (E) Shannon diversity. Significance for (C)–(E) was determined using a Kruskal-Wallis non-parametric ANOVA (p values inset at the bottom of each panel) with Dunn’s multiple comparison. ∗∗p < 0.01, ∗∗∗∗ = p < 0.0001.
(F) Bubble plots of bacterial genera with >1% average abundance across the entire study population. The size of each circle indicates the average relative abundance for each taxa, and the color of each circle denotes bacterial phyla.