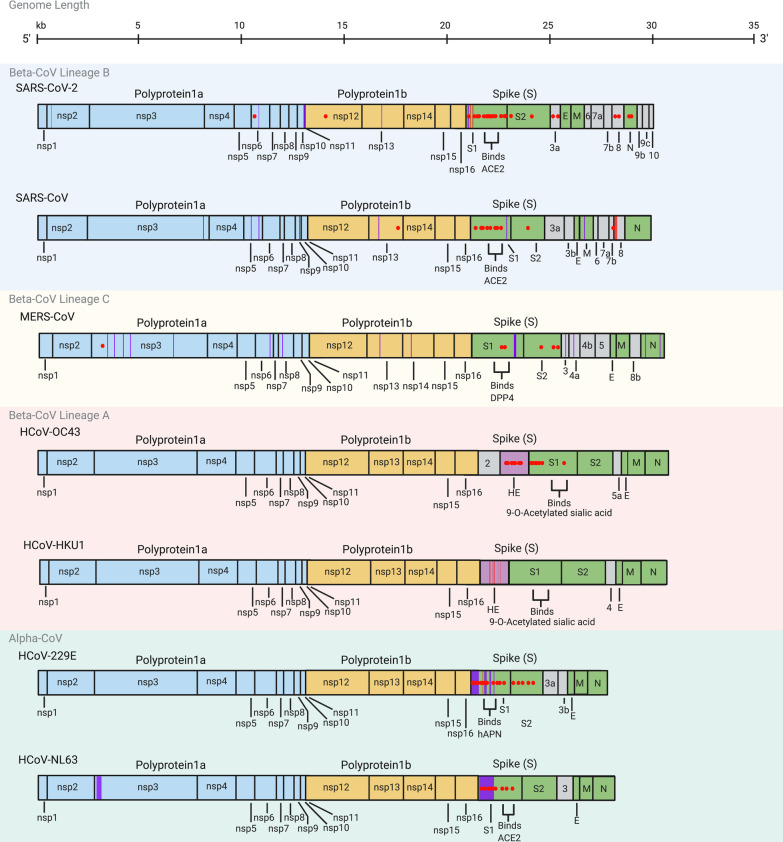
Fig. 2.
Mutations identified in human coronaviruses. Red dots within the genomes correspond to specific amino acid residues that have been strongly positively selected for such that a specific mutation has become dominant in the region where it emerged [74, 78, 83–91, 94–96, 99–101, 104, 111, 116, 117, 121, 123–125, 129, 131, 132, 135, 138–140, 146, 151–154, 158, 162, 278, 284–293]. Genomic regions highlighted by red bars correspond to deletions that have been selected for, while purple bars correspond to regions with significant polymorphisms within a CoV species. Beta-CoV Lineage B (Sarbecovirus) is represented within the blue shaded area, beta-CoV Lineage C (Merbecovirus) is represented within the yellow shaded area, beta-CoV Lineage A (Embecovirus) is represented within the red shaded area, and alpha-CoVs are represented within the green shaded area. Genome length in kilobases (kb) is noted on top. See Table 2 for more details