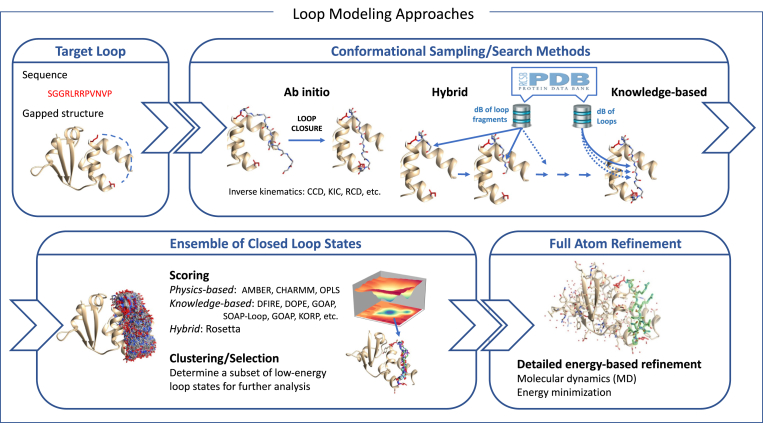
Fig. 3.
Schematic workflow of a prototype loop modeling protocol. It involves several stages. Given a three-dimensional model of a protein and the loop sequence (required if the loop is missing in the input structure), the first stage generates possible states of the loop. This can be based on ab initio sampling, searches in structural repositories, or a combination of both approaches. When the number of sampled states is large, scoring and clustering methods are useful to select a reduced number of the most likely loop models. These models can be subsequently refined at the all-atom level, considering the flexibility of the surrounding parts of the protein, and possibly including a model of the solvent.