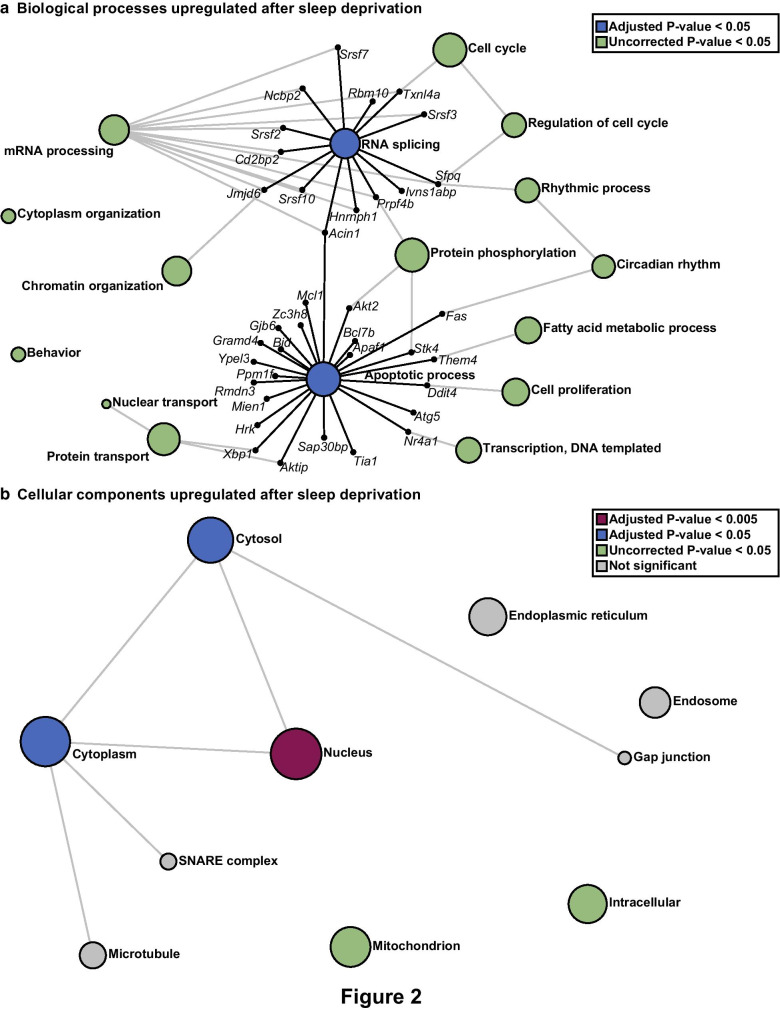
Fig. 2.
Distinct biological processes and cellular components are enriched for genes upregulated after acute sleep deprivation. a Using Network Analyst software and the PANTHER: Biological Processes (BP) classification to perform overrepresentation analysis (ORA), pathways enriched for upregulated genes were identified. RNA splicing and Apoptotic Process were the most significantly enriched networks (Adjusted P-value = 0.025). These top networks have been expanded to show the genes that are involved and upregulated after sleep deprivation. b Using Network Analyst software and the PANTHER: Cellular Components (CC) classification to perform ORA we identified cellular components enriched for genes upregulated after sleep deprivation. The nucleus is the most significantly enriched (Adjusted P-value = 1.74 × 10–9). The size of each node represents the number of hits from the inputted gene list