Figure 5.
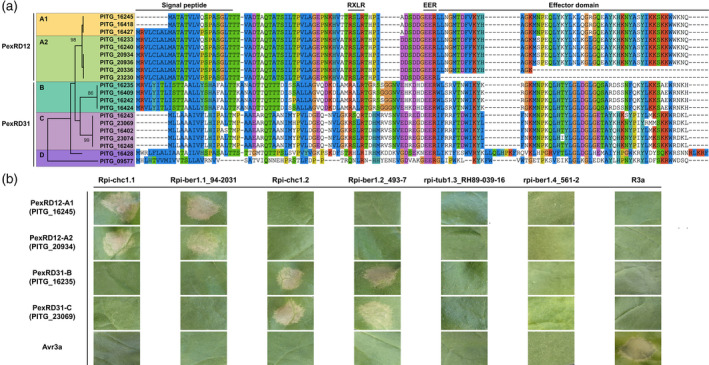
Rpi‐chc1 alleles show non‐overlapping recognition of the PexRD12/31 effector superfamily.
(a) Twenty members of the PexRD12/31 superfamily were found in P. infestans isolate T30‐4. In the amino acid sequence, we can distinguish a signal peptide in the N‐terminus, the conserved RXLR‐EER motifs in the center, and the effector domain in the C‐terminus. Some PexRD12/31 family members differed at the nucleotide level but were identical at the protein level (PITG_16245 = PITG_16418; PITG_16233 = PITG_16240; PITG_20934 = PITG_20936; PITG_16409 = PITG_16424). The phylogenetic analysis of the complete protein sequences led to the identification of five clades. This analysis was performed in mega x by using the maximum likelihood method based on the JTT matrix‐based model. The tree with the highest log‐likelihood (−766) is shown. The bootstrapping values, which indicate the percentage of trees that had the particular branch, are shown in each branch. In the protein alignment: blue, hydrophobic residues (A, I, L, M, F, W, V, and C); red, positively charged residues (K and R); magenta, negatively charged residues (E and D); green, polar residues (N, Q, S, and T); pink, cysteine residues (C); orange, glycine residues (G); yellow, proline residues (P); cyan, aromatic residues (H and Y); and white, unconserved residues or gaps. (b) Different Rpi‐chc1 allelic variants were co‐agroinfiltrated in N. benthamiana with a member from each PexRD12/31 clade. While variants from clade 1 recognize both PexRD12‐A1 and ‐A2 clades, Rpi‐chc1 variants from clade 2 recognize PexRD31‐B and ‐C. Receptors from clades 3 and 4 do not recognize any PexRD12/31 effector. A mix of R3a and Avr3a was used as a positive control.
