Figure 6.
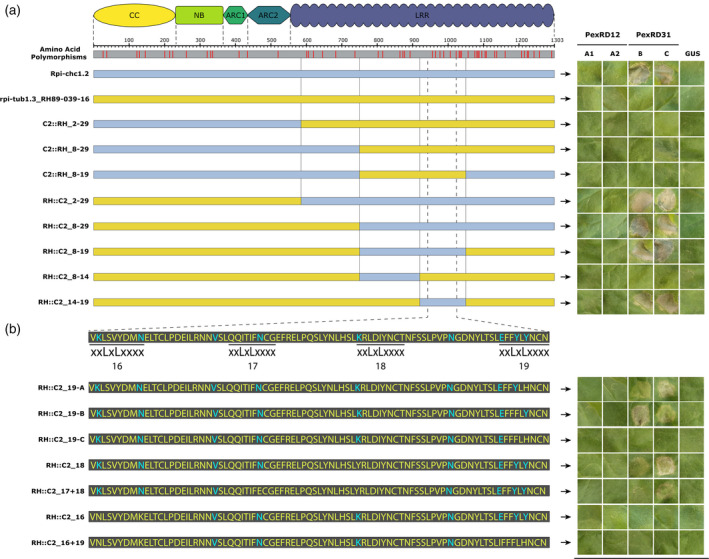
Domain exchanges between Rpi‐chc1.2 and rpi‐tub1.3_RH89‐039‐16.
(a) The positions of SAPs and the corresponding protein domains are indicated on top. Rpi‐chc1.2 and rpi‐tub1.3_RH89‐039‐16 are represented as light blue and yellow bars, respectively. Below, the domain exchanges are shown. The chimeric constructs were co‐agroinfiltrated with the PexRD12/31 effectors in N. benthamiana leaves. After 4 days, the HR was visible and recorded. Experiments were repeated three times with 12 inoculation sites each time. A representative leaf for the response of each chimeric construct is shown in the right panel. GUS was used as a negative control. It is concluded that the exchange of the complete LRR domain led to recognition of PexRD31. With the final construct, RH::C2_14‐19, the exchange of only nine amino acids led to the activation of the rpi‐tub1.3_RH89‐036‐16 protein. (b) Seven new modified receptors were derived from RH::C2_14‐19 in order to pinpoint the amino acids involved in the Rpi‐chc1.2 recognition specificity. SAPs present in Rpi‐chc1.2 are highlighted in blue font. Most of the SAPs are located in the solvent‐exposed xxLxLxxxx motifs of LRRs 16–19. The chimeric constructs were co‐agroinfiltrated with the PexRD12/31 effectors in N. benthamiana leaves. A representative leaf for the response of each chimeric construct is shown in the right panel. Experiments were repeated three times with 12 inoculation sites each time. GUS was used as a negative control. The modification of the tested residues of the Rpi‐chc1.2 solvent‐exposed specific amino acids (blue) for the corresponding amino acid present in rpi‐tub1.3_RH89‐039‐16 (yellow) led to the partial or complete loss of effector recognition.
