FIGURE 1.
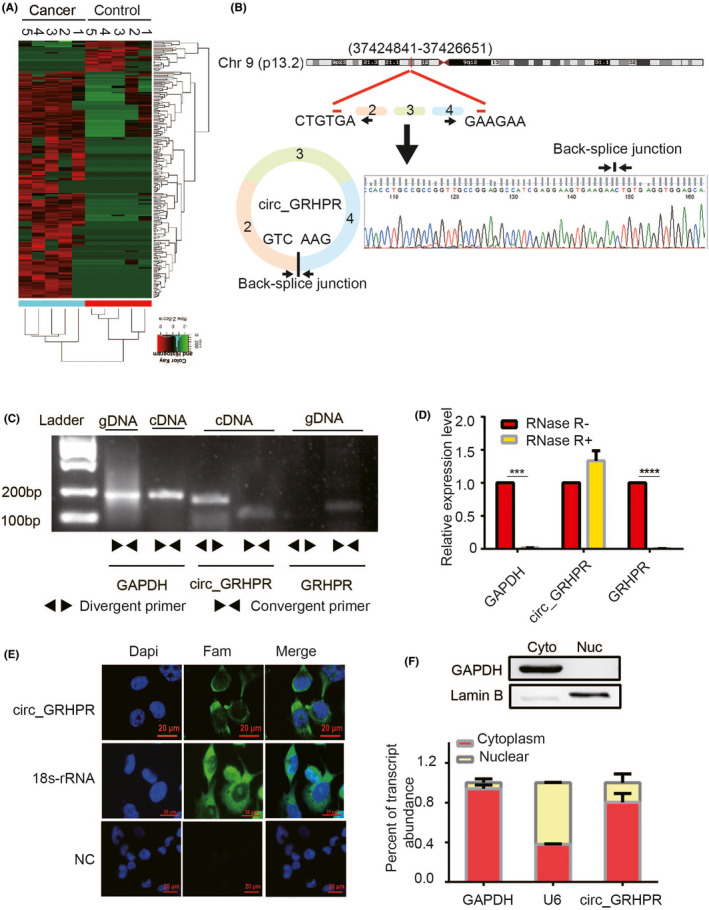
Validation and stability analysis of circ_GRHPR in non‐small cell lung cancer (NSCLC). (A) Heat maps of differential expression of circular RNAs in lung cancer patients (n = 5) and controls (n = 5) are shown (red indicates upregulated expression; green indicates downregulated expression). (B) The schematic diagram shows the structure of circ_GRHPR via the circularisation of exons 2, 3, and 4 from GRHPR in Chr9; Sanger sequencing of the RT‐PCR products of circ_GRHPR is shown on the right. (C) circ_GRHPR and GRHPR were amplified by cDNA and gDNA of H1299 cells with divergent primers and convergent primers, respectively. (D) The expression of circ_GRHPR and GRHPR in H1299 cells were detected by qRT‐PCR in the presence or absence of RNase R. GAPDH was used as an internal control. (E) Subcellular localisation of circ_GRHPR was detected by FISH, 18S‐rRNA was used as the positive control, and a negative control probe was used as the negative control in NSCLC cells. (F) Location of circ_GRHPR in NSCLC cells. (Left) GAPDH and U6 were applied as positive controls in the cytoplasm and nucleus, respectively. Western blotting was used to detect the partitioning between the nucleus and cytoplasm. Data are shown as the mean ± SD, p < 0.05 was considered statistically significant (*p < 0.05/**p < 0.01/***p < 0.001)
