Extended Data Fig. 6. In vivo overexpression of SLC2A3 increased vascular leak.
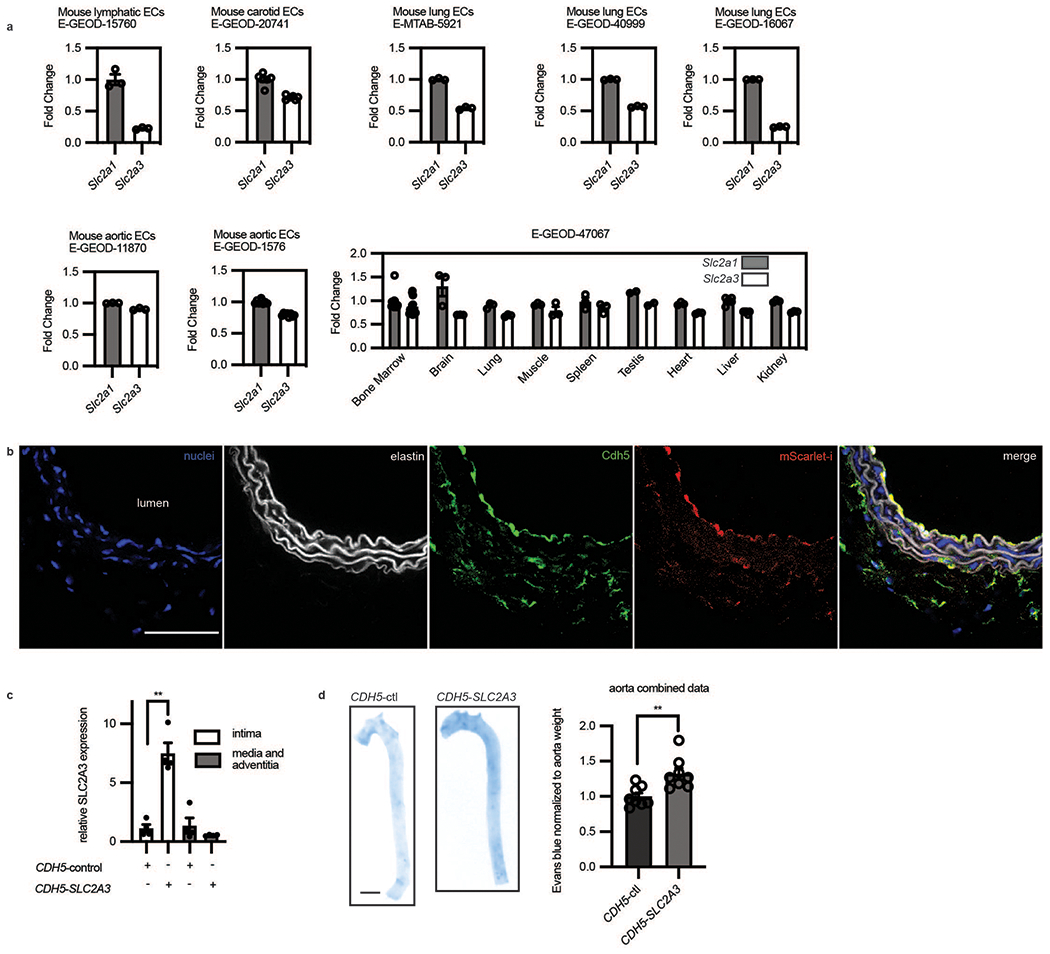
a, GEO datasets of primary endothelial mouse cells comparing Slc2a1 and Slc2a3 from different organs. Fold change indicates that data are normalized to Slc2a1 except for E-GEOD-47067 which was normalized to average of Slc2a1 in all tissues. Only controls of experiments in the GEO datasets were used in these analyses. (n = 3 independent biological replicates for each condition for E-GEOD-15760; n = 6 independent biological replicates for each condition for E-GEOD-20741; n = 3 independent biological replicates for each condition for E-MTAB-5921; n = 3 independent biological replicates for each condition for E-GEOD-40999; n = 3 independent biological replicates for each condition for E-GEOD-11870; n = 3 independent biological replicates for each condition for E-GEOD-1576; n = 3 independent biological replicates for condition in each organ for E-GEOD-47067) b, Immunofluorescence of an arterial section demonstrating enhanced mScarlet-i fluorescence (red) colocalized with ve-cadh (green) which marks the endothelium next to the lumen (yellow, merge) (Scale bar, 500 μm, representative of n = 4 mice). c, SLC2A3 expressing plasmid uses an endothelial specific CDH5 promoter. SLC2A3 is detected by qRT-PCR of the intima of the mouse aorta, and far less than in the media and adventitia. (n = 4 independent biological replicates, p = 0.0030) d, Evans blue (OD 620 nm) quantification of mouse aortas, normalized by weight. (n = 8 independent biological replicates for each condition, p = 0.0050). Statistical significance was determined by two-sided Welch’s t-test. All error bars are SEM.
