Fig 1. Overview of the AI approach used to obtain a model for the classification of a sequence as an aptamer.
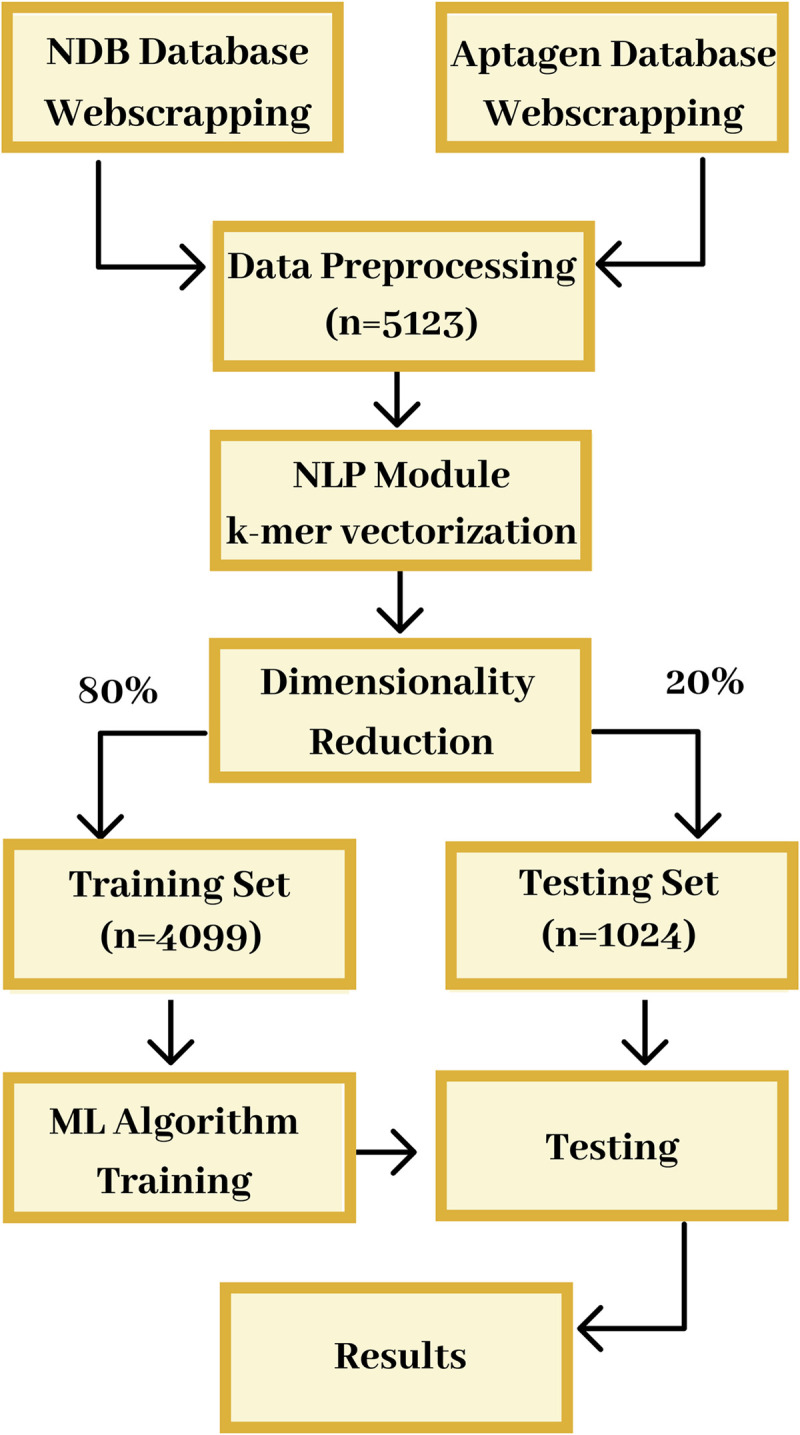
It included the extraction of nucleotide sequences from the Nucleic Acid Database (NDB) and Aptagen. The sequences were converted into 6-mer vectors using the NLP modules. Out of the 5,123 vectors created, only the top 2.5% were selected for modeling, in the reduction of dimensionality module. Then the data was split into a training set (80% of the data, n = 4,099) and test set (20% of the data, n = 1,024). Because of data imbalance in the training set, the underrepresented samples were weighted highly. ML algorithms were trained to develop the models using the selected features. The developed models were tested using cross-validation and validated using the test sets. Fig 1 is also the Graphical Abstract.
