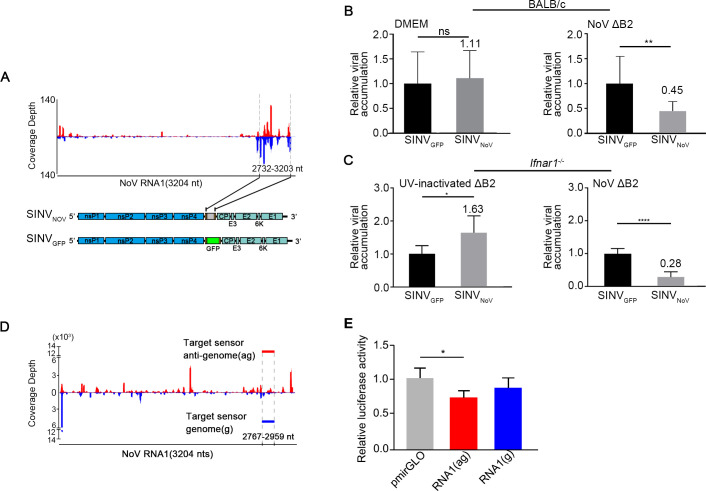
Fig 5. The viral siRNAs of NoV inducing homology-dependent viral RNA degradation.
A. The genomic structure of a recombinant SINV (SINVNoV) carrying a segment of NoV RNA1 known to be targeted by high densities of vsiRNAs in NoVΔB2-infected mice shown at the top. Also shown below was the genomic structure of SINVGFP introduced above. B. In vivo genomic RNA levels of SINVNoV and SINVGFP one day after challenge inoculation of NoVΔB2-infected (right) or DMEM mock-infected (left) BALB/c suckling mice (n = 7 per DMEM group, n = 10 per NoVΔB2 group). The viral genomic RNA accumulation level was determined by RT-qPCR amplification of the viral nsP2 coding region and normalized by endogenous actin mRNA with SINVGFP level set as 1. Error bars represent SD. ** indicates p<0.01 (Student’s t-test). C. In vivo genomic RNA levels of SINVNoV and SINVGFP measured as in (B) one day after challenge inoculation of Ifnar1-/- suckling mice (n = 5~7 per group) inoculated two days earlier with live (right) or UV-inactivated (left) NoVΔB2. Error bars represent SD. * indicates <0.05, **** indicates p<0.0001 (Student’s t-test). The viral RNA accumulation of SINVGFP was set as 1. D. Diagram showing the genomic position of the 200-nt NoV sequence in the genome (g, blue) or anti-genome (ag, red) sense inserted into the 3′ untranslated region (UTR) of firefly luciferase reporter mRNA, to be targeted respectively by antisense and sense vsiRNAs extracted from VSR-B2 immuno-precipitants of NoV-infected C57BL/6 suckling mice. E. Relative luciferase activity of the control and chimeric reporter constructs with a 3’-UTR containing a NoV sequence targeted by the positive- or negative-strand vsiRNAs sequestered in VSR-B2 complex. Error bars indicate standard deviation of three replicates. Error bars represent SD. * indicates p<0.05 (Student’s t-test).