FIGURE 3: The forkhead motif regulates Foxp3 binding but not accessibility of Foxp3 targets.
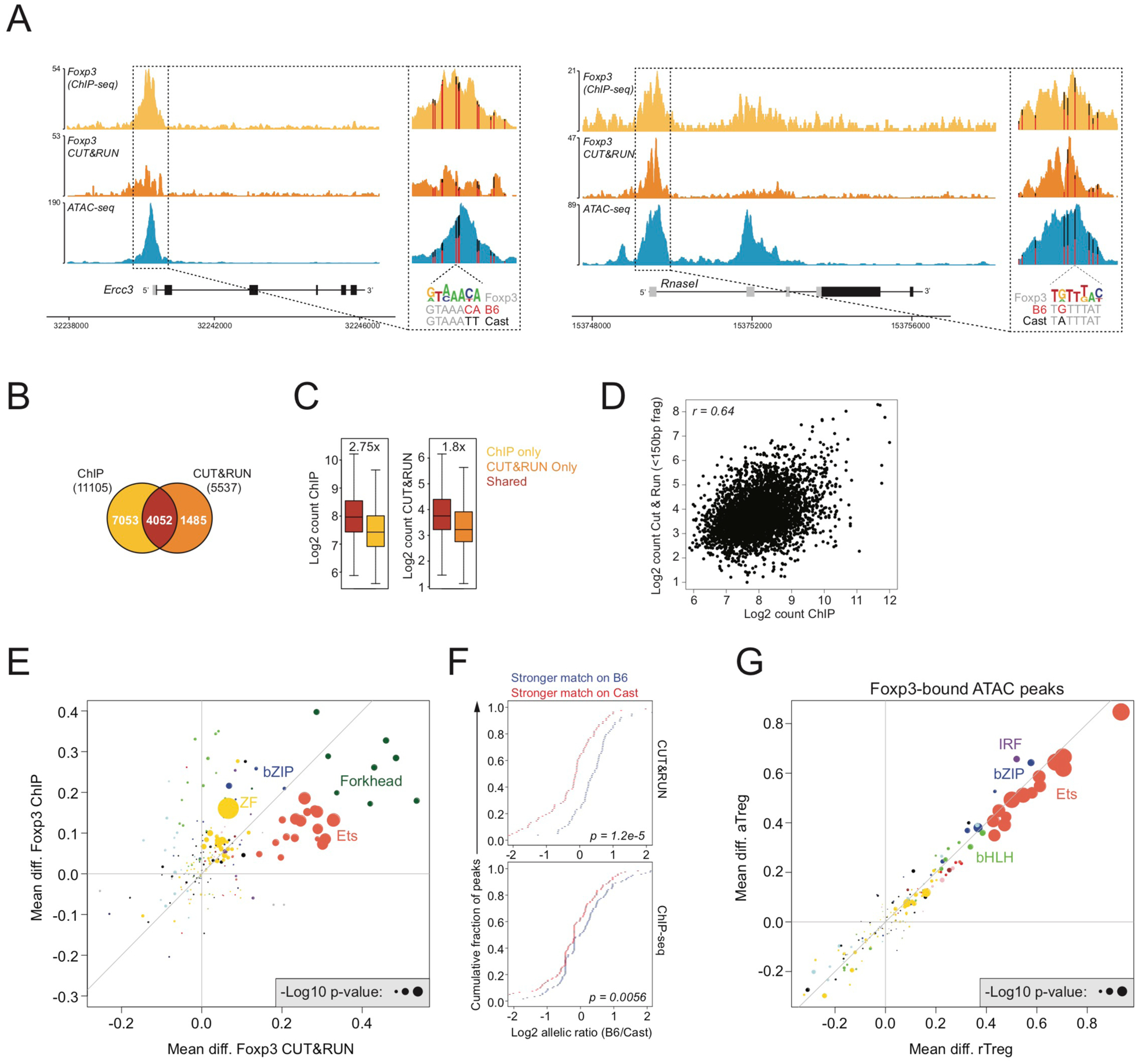
(A): Foxp3 ChIP-seq, Foxp3 CUT&RUN, and ATAC-seq (rTreg) tracks. Vertical bars mark the position of genetic variants, with color indicating the ratio of sequencing reads derived from each allele. A Foxp3 motif variant is highlighted.
(B): Overlap between Foxp3 binding sites identified by ChIP or CUT&RUN in bulk Treg cells from pooled spleen and lymph nodes of Foxp3DTR-GFP B6/Cast F1 mice.
(C): Read counts at Foxp3 binding sites identified by ChIP-seq, CUT&RUN, or by both techniques.
(D): Pearson correlation between Foxp3 binding intensity measured by ChIP and CUT&RUN for sites detected by both techniques.
(E): Effect of genetic variation in TF binding motifs on allelic bias in Foxp3 binding, measured by ChIP or CUT&RUN. See Figure S1.
(F): Effect of Foxp3 motif variation on allelic bias in Foxp3 binding by ChIP (bottom) or CUT&RUN (top).
(G): Effect of genetic variation in TF binding motifs on allelic bias in chromatin accessibility (ATAC-seq) of Foxp3-bound sites.
