Figure 2.
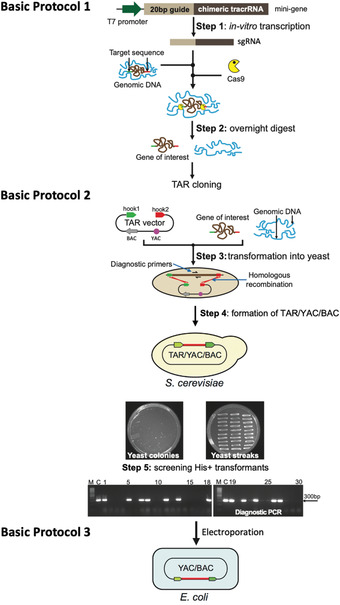
Diagram of the updated TAR‐CRISPR/Cas9 method for selective isolation of a gene/locus of interest from complex genomes. Basic Protocol 1, Step 1: The mini‐gene is in vitro transcribed to make sgRNA. Prepared genomic DNA is then cleaved in vitro at positions close to the 5′ and 3′ ends of a gene/locus by specifically designed Cas9‐sgRNA complexes. Step 2: A digestion mix containing sgRNAs, Cas9, and genomic DNA containing a gene of interest is prepared and left to digest overnight. Basic Protocol 2, Step 3: Prepared yeast spheroplasts are transformed with CRISPR/Cas9‐treated genomic DNA along with the linearized TAR vector containing BAC and YAC cassettes and the hooks corresponding to the 5′ and 3′ ends of a gene of interest (hook 1 in green and hook 2 in red). Step 4: Homologous recombination between the targeting sequences in the vector and the targeted genomic DNA fragment ends leads to the establishment of a circular TAR/YAC/BAC molecule. A representative His‐ plate with yeast His+ transformants after 5‐7 days of growth obtained by TAR cloning of the human NBS1 gene from total genomic DNA is shown, together with a plate with 30 streaked randomly chosen His+ transformants after 1 day of growth. Step 5: Analysis of the streaked His+ transformants by PCR for the presence of the gene of interest using a pair of diagnostic primers. An example of the screening of transformants harboring the human NSB1 gene is shown. M‐GeneRuler 1 kb DNA Ladder (Thermo Fisher Scientific, cat. no. SM0311); C‐control PCR with human genomic DNA. Basic Protocol 3: YAC/BAC DNAs containing a gene/region of interest are prepared in agarose plugs and then transferred from yeast to E. coli cells by electroporation for further BAC DNA isolation, sequencing, and analysis.
