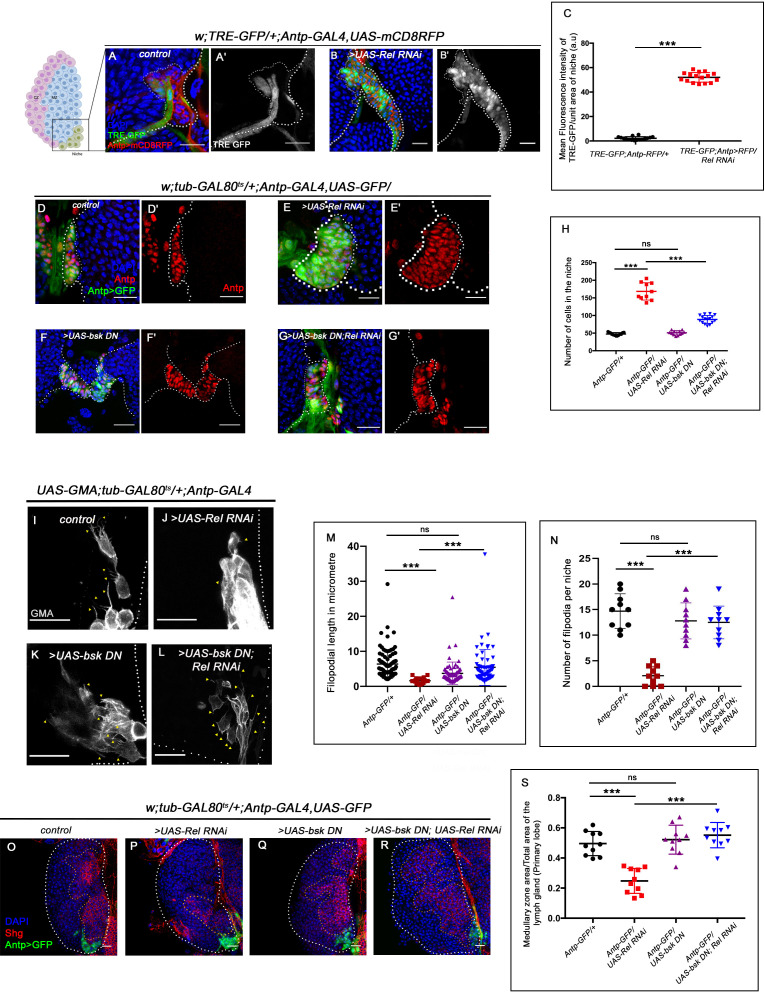
Figure 5. Loss of Relish from the niche activated JNK causing niche hyperplasia.
The genotypes are mentioned in relevant panels. Scale bar: 20 μm. (A–B') Upregulation of JNK signaling visualized by its reporter TRE-GFP (green) in Relish knockdown (B–B') compared with WT niche (A–A'). (C) Statistical analysis of fluorescence intensity (A–B') revealed a significant increase in TRE-GFP levels compared to control (n=15, p-value=4.2 × 10−19, two-tailed Student’s t-test). (D–G') Upon niche-specific simultaneous knockdown of Rel and JNK, the niche hyperplasia observed upon loss of Relish (E–E') is rescued (G–G') and is comparable to control (D–D') whereas loss of bsk from the niche does not alter niche cell number (F–F'). (H) Statistical analysis of the data in (D–G') (n=10, p-value=5.6 × 10−8 for control versus Rel RNAi, p-value=8.0 × 10−7 for bsk DN; Rel RNAi versus Rel RNAi, p-value=0.10 control versus for bsk DN; two-tailed unpaired Student's t-test). (I–N) Cellular filopodia from the niche cells in Rel loss of function is found to be smaller in length and fewer in numbers (J and M–N). Simultaneous loss of both JNK using bsk DN and Relish (L and M–N) rescued the stunted, scanty filopodia to control state (I and M–N), whereas loss of JNK did not affect filopodia formation (K and M–N). (M–N) Statistical analysis of the data in (I–L) (Filopodia number: n=10, p=6.96 × 10−8 for control versus Rel RNAi, p-value=8.11 × 10−7 for bsk DN; Rel RNAi versus Rel RNAi, p-value=0.153 for bsk DN versus control. Filopodia length: n=6, p-value=2.78 × 10−16 for control versus Rel RNAi, p-value=1.84 × 10−6 for bsk DN; Rel RNAi versus Rel RNAi, p-value=0.22 for bsk DN vs control; two-tailed unpaired Student’s t-test). (O–R) Knocking down JNK function from the niche did not have any effect on progenitors (visualized by Shg) (Q). Downregulating bsk function in Rel loss genetic background was able to restore the reduction in prohemocyte pool (R) observed in Relish loss (P) scenario in comparison to control (O). (S) Statistical analysis of the data in (O–R) (n=10, p-value=2.26 × 10−6 for control versus Rel RNAi, p-value=1.94 × 10−7 for bsk DN; Rel RNAi versus Rel RNAi, p-value=0.521 for control versus bsk DN; two-tailed unpaired Student's t-test) The white dotted line marks whole of the lymph gland in all cases and niches in (A–G'). Yellow dotted lines mark the progenitor zone in (O–R). In all panels, age of the larvae is 96 hr AEH. The nuclei are marked with DAPI (blue). Individual dots represent biological replicates. Error bar: standard deviation (SD). Data are mean ± SD. *p<0.05, **p<0.01, and ***p<0.001.