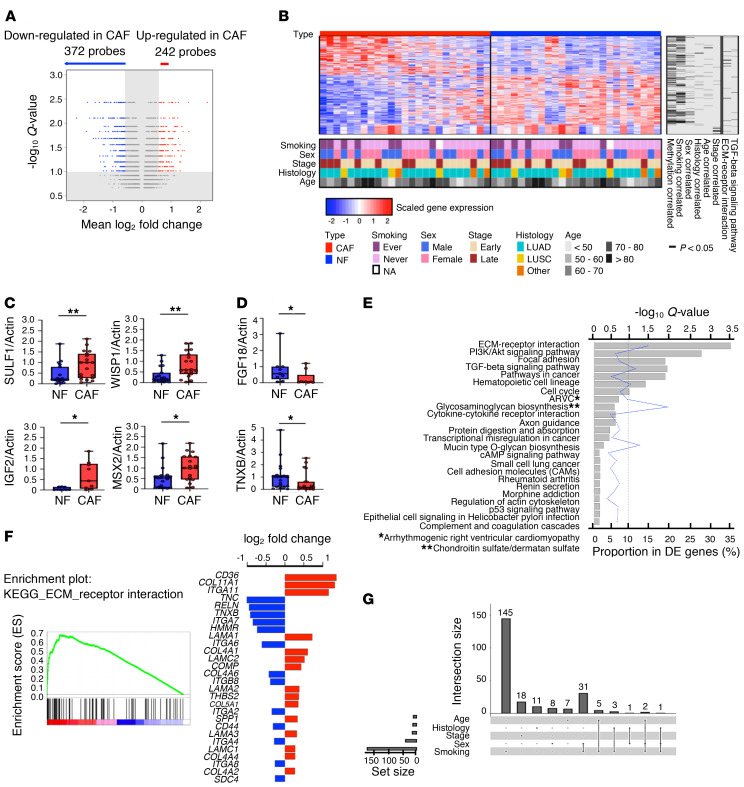
Figure 2. Identification of differential gene expression between lung NFs and CAFs.
(A) Volcano plot of the differentially expressed (DE) probes between NFs and CAFs. A probe is significant if fold change > 1.5 and Q < 0.1. (B) Heatmap of 614 DE probes on 25 pairs of primarily cultured NF/CAF samples from NSCLC patients. The standardized log2 expression values are displayed. Rows represent probes and columns represent samples. The clinical characteristics are encoded in the bottom and the significantly (P < 0.05 by Welch’s t test; for age, Pearson’s correlation coefficient was tested using the t test) correlated DNA methylation data, clinical variables, and related pathways are indicated in the right columns. (C and D) Analysis of DE genes, upregulated (C) or downregulated (D) in CAFs, in NF/CAF pairs (n = 19 or n = 9 per gene) by quantitative real-time PCR (qPCR). Actin was used as the internal control. Data presented as mean ± SD; symbols represent individual samples. *P < 0.05, **P < 0.01 by Mann-Whitney U test. Box plots display the median, first and third quartiles with whiskers as maximum and minimum values. (E) Using DAVID analysis, 7 KEGG pathways were significantly enriched in the DE genes at the false discovery rate of 0.1. (F) GSEA enrichment plot for ECM-receptor interaction pathway in comparing CAFs (red) to NFs (blue). The log2 fold changes of the core genes are shown in the right panel. (G) UpSet plot for the intersections among the 5 sets of the DE probes correlated (P < 0.05 by Welch’s t test; for age, Pearson’s correlation coefficient was tested using the t test) with tumor histology, stage, age, sex, and patients’ smoking status. The set of smoking-correlated DE probes overwhelmingly outnumbered other variables.